Quantitative cardiac phosphoproteomics profiling during ischemia-reperfusion in an immature swine model
- PMID: 28455290
- PMCID: PMC5538860
- DOI: 10.1152/ajpheart.00842.2016
Quantitative cardiac phosphoproteomics profiling during ischemia-reperfusion in an immature swine model
Abstract
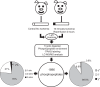
Ischemia-reperfusion (I/R) results in altered metabolic and molecular responses, and phosphorylation is one of the most noted regulatory mechanisms mediating signaling mechanisms during physiological stresses. To expand our knowledge of the potential phosphoproteomic changes in the myocardium during I/R, we used Isobaric Tags for Relative and Absolute Quantitation-based analyses in left ventricular samples obtained from porcine hearts under control or I/R conditions. The data are available via ProteomeXchange with identifier PXD006066. We identified 1,896 phosphopeptides within left ventricular control and I/R porcine samples. Significant differential phosphorylation between control and I/R groups was discovered in 111 phosphopeptides from 86 proteins. Analysis of the phosphopeptides using Motif-x identified five motifs: (..R..S..), (..SP..), (..S.S..), (..S…S..), and (..S.T..). Semiquantitative immunoblots confirmed site location and directional changes in phosphorylation for phospholamban and pyruvate dehydrogenase E1, two proteins known to be altered by I/R and identified by this study. Novel phosphorylation sites associated with I/R were also identified. Functional characterization of the phosphopeptides identified by our methodology could expand our understanding of the signaling mechanisms involved during I/R damage in the heart as well as identify new areas to target therapeutic strategies.NEW & NOTEWORTHY We used Isobaric Tags for Relative and Absolute Quantitation technology to investigate the phosphoproteomic changes that occur in cardiac tissue under ischemia-reperfusion conditions. The results of this study provide an extensive catalog of phosphoproteins, both predicted and novel, associated with ischemia-reperfusion, thereby identifying new pathways for investigation.
Keywords: Isobaric Tags for Relative and Absolute Quantitation; ischemia-reperfusion; phosphoproteomics.
Copyright © 2017 the American Physiological Society.
Figures




References
-
- Ambrus A, Wang J, Mizsei R, Zambo Z, Torocsik B, Jordan F, Adam-Vizi V. Structural alterations induced by ten disease-causing mutations of human dihydrolipoamide dehydrogenase analyzed by hydrogen/deuterium-exchange mass spectrometry: Implications for the structural basis of E3 deficiency. Biochim Biophys Acta 1862: 2098–2109, 2016. doi:10.1016/j.bbadis.2016.08.013. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

