Three Cdk1 sites in the kinesin-5 Cin8 catalytic domain coordinate motor localization and activity during anaphase
- PMID: 28455557
- PMCID: PMC11107736
- DOI: 10.1007/s00018-017-2523-z
Three Cdk1 sites in the kinesin-5 Cin8 catalytic domain coordinate motor localization and activity during anaphase
Abstract
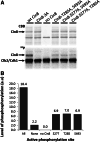
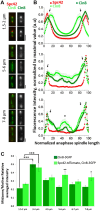
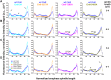
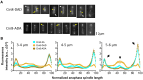
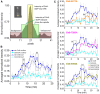
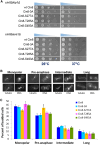
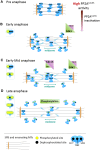
The bipolar kinesin-5 motors perform essential functions in mitotic spindle dynamics. We previously demonstrated that phosphorylation of at least one of the Cdk1 sites in the catalytic domain of the Saccharomyces cerevisiae kinesin-5 Cin8 (S277, T285, S493) regulates its localization to the anaphase spindle. The contribution of these three sites to phospho-regulation of Cin8, as well as the timing of such contributions, remains unknown. Here, we examined the function and spindle localization of phospho-deficient (serine/threonine to alanine) and phospho-mimic (serine/threonine to aspartic acid) Cin8 mutants. In vitro, the three Cdk1 sites undergo phosphorylation by Clb2-Cdk1. In cells, phosphorylation of Cin8 affects two aspects of its localization to the anaphase spindle, translocation from the spindle-pole bodies (SPBs) region to spindle microtubules (MTs) and the midzone, and detachment from the mitotic spindle. We found that phosphorylation of S277 is essential for the translocation of Cin8 from SPBs to spindle MTs and the subsequent detachment from the spindle. Phosphorylation of T285 mainly affects the detachment of Cin8 from spindle MTs during anaphase, while phosphorylation at S493 affects both the translocation of Cin8 from SPBs to the spindle and detachment from the spindle. Only S493 phosphorylation affected the anaphase spindle elongation rate. We conclude that each phosphorylation site plays a unique role in regulating Cin8 functions and postulate a model in which the timing and extent of phosphorylation of the three sites orchestrates the anaphase function of Cin8.
Keywords: Anaphase B; Cin8; Kinesin-5; Microtubules; Mitosis; Phospho-regulation.
Figures

Similar articles
-
Bidirectional motility of kinesin-5 motor proteins: structural determinants, cumulative functions and physiological roles.Cell Mol Life Sci. 2018 May;75(10):1757-1771. doi: 10.1007/s00018-018-2754-7. Epub 2018 Feb 3. Cell Mol Life Sci. 2018. PMID: 29397398 Free PMC article. Review.
-
Synthetic-Evolution Reveals Narrow Paths to Regulation of the Saccharomyces cerevisiae Mitotic Kinesin-5 Cin8.Int J Biol Sci. 2019 May 2;15(6):1125-1138. doi: 10.7150/ijbs.30543. eCollection 2019. Int J Biol Sci. 2019. PMID: 31223274 Free PMC article.
-
Phospho-regulation of kinesin-5 during anaphase spindle elongation.J Cell Sci. 2011 Mar 15;124(Pt 6):873-8. doi: 10.1242/jcs.077396. J Cell Sci. 2011. PMID: 21378308 Free PMC article.
-
Phosphorylation-dependent protein interactions at the spindle midzone mediate cell cycle regulation of spindle elongation.Dev Cell. 2009 Aug;17(2):244-56. doi: 10.1016/j.devcel.2009.06.011. Dev Cell. 2009. PMID: 19686685
-
Factors that Control Mitotic Spindle Dynamics.Adv Exp Med Biol. 2017;925:89-101. doi: 10.1007/5584_2016_74. Adv Exp Med Biol. 2017. PMID: 27722958 Review.
Cited by
-
Bidirectional motility of kinesin-5 motor proteins: structural determinants, cumulative functions and physiological roles.Cell Mol Life Sci. 2018 May;75(10):1757-1771. doi: 10.1007/s00018-018-2754-7. Epub 2018 Feb 3. Cell Mol Life Sci. 2018. PMID: 29397398 Free PMC article. Review.
-
The microtubule plus-end tracking protein Bik1 is required for chromosome congression.Mol Biol Cell. 2022 May 1;33(5):br7. doi: 10.1091/mbc.E21-10-0500. Epub 2022 Mar 2. Mol Biol Cell. 2022. PMID: 35235370 Free PMC article.
-
Intracellular functions and motile properties of bi-directional kinesin-5 Cin8 are regulated by neck linker docking.Elife. 2021 Aug 13;10:e71036. doi: 10.7554/eLife.71036. Elife. 2021. PMID: 34387192 Free PMC article.
-
PTP-3 phosphatase promotes intramolecular folding of SYD-2 to inactivate kinesin-3 UNC-104 in neurons.Mol Biol Cell. 2020 Dec 15;31(26):2932-2947. doi: 10.1091/mbc.E19-10-0591. Epub 2020 Nov 4. Mol Biol Cell. 2020. PMID: 33147118 Free PMC article.
-
Synthetic-Evolution Reveals Narrow Paths to Regulation of the Saccharomyces cerevisiae Mitotic Kinesin-5 Cin8.Int J Biol Sci. 2019 May 2;15(6):1125-1138. doi: 10.7150/ijbs.30543. eCollection 2019. Int J Biol Sci. 2019. PMID: 31223274 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

