Differences in gut microbial composition correlate with regional brain volumes in irritable bowel syndrome
- PMID: 28457228
- PMCID: PMC5410709
- DOI: 10.1186/s40168-017-0260-z
Differences in gut microbial composition correlate with regional brain volumes in irritable bowel syndrome
Abstract
Background: Preclinical and clinical evidence supports the concept of bidirectional brain-gut microbiome interactions. We aimed to determine if subgroups of irritable bowel syndrome (IBS) subjects can be identified based on differences in gut microbial composition, and if there are correlations between gut microbial measures and structural brain signatures in IBS.
Methods: Behavioral measures, stool samples, and structural brain images were collected from 29 adult IBS and 23 healthy control subjects (HCs). 16S ribosomal RNA (rRNA) gene sequencing was used to profile stool microbial communities, and various multivariate analysis approaches were used to quantitate microbial composition, abundance, and diversity. The metagenomic content of samples was inferred from 16S rRNA gene sequence data using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt). T1-weighted brain images were acquired on a Siemens Allegra 3T scanner, and morphological measures were computed for 165 brain regions.
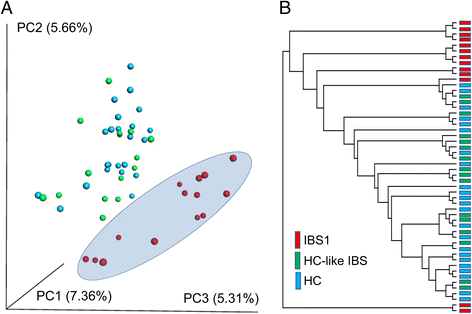
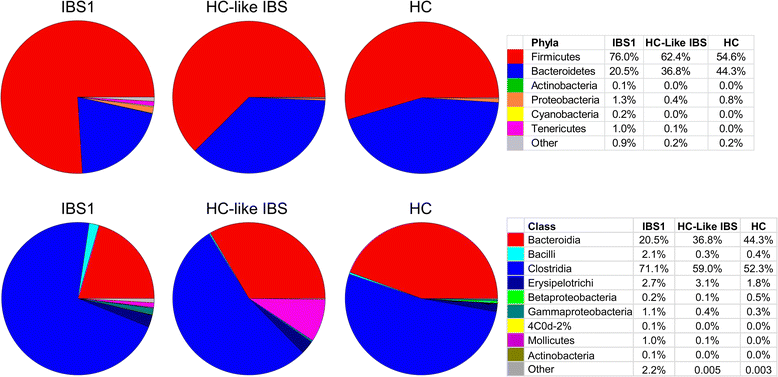
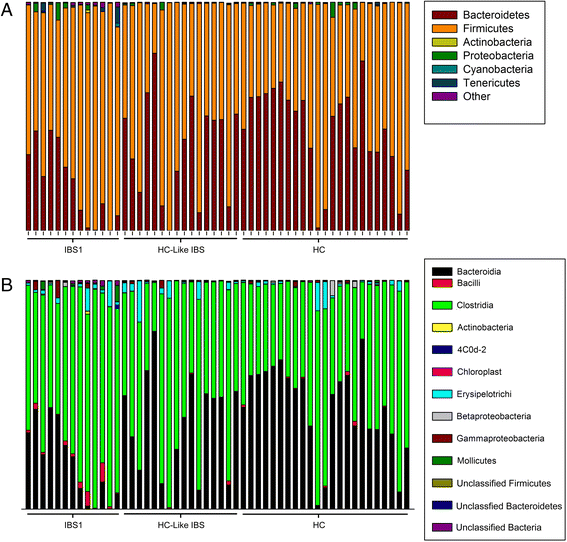
Results: Using unweighted Unifrac distances with hierarchical clustering on microbial data, samples were clustered into two IBS subgroups within the IBS population (IBS1 (n = 13) and HC-like IBS (n = 16)) and HCs (n = 23) (AUROC = 0.96, sensitivity 0.95, specificity 0.67). A Random Forest classifier provided further support for the differentiation of IBS1 and HC groups. Microbes belonging to the genera Faecalibacterium, Blautia, and Bacteroides contributed to this subclassification. Clinical features distinguishing the groups included a history of early life trauma and duration of symptoms (greater in IBS1), but not self-reported bowel habits, anxiety, depression, or medication use. Gut microbial composition correlated with structural measures of brain regions including sensory- and salience-related regions, and with a history of early life trauma.
Conclusions: The results confirm previous reports of gut microbiome-based IBS subgroups and identify for the first time brain structural alterations associated with these subgroups. They provide preliminary evidence for the involvement of specific microbes and their predicted metabolites in these correlations.
Keywords: Bacteroidetes; Brain-gut-microbiome axis; Firmicutes; Irritable bowel syndrome; Metagenome.
Figures


References
-
- Drossman DA. Functional gastrointestinal disorders: history, pathophysiology, clinical features and Rome IV. Gastroenterol. 2016. Feb 19. [Epub ahead of print] - PubMed

