Gli1+ Mesenchymal Stromal Cells Are a Key Driver of Bone Marrow Fibrosis and an Important Cellular Therapeutic Target
- PMID: 28457748
- PMCID: PMC6485654
- DOI: 10.1016/j.stem.2017.03.008
Gli1+ Mesenchymal Stromal Cells Are a Key Driver of Bone Marrow Fibrosis and an Important Cellular Therapeutic Target
Erratum in
-
Gli1+ Mesenchymal Stromal Cells Are a Key Driver of Bone Marrow Fibrosis and an Important Cellular Therapeutic Target.Cell Stem Cell. 2018 Aug 2;23(2):308-309. doi: 10.1016/j.stem.2018.07.006. Cell Stem Cell. 2018. PMID: 30075132 No abstract available.
Abstract
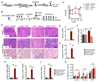
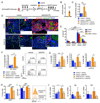
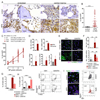
Bone marrow fibrosis (BMF) develops in various hematological and non-hematological conditions and is a central pathological feature of myelofibrosis. Effective cell-targeted therapeutics are needed, but the cellular origin of BMF remains elusive. Here, we show using genetic fate tracing in two murine models of BMF that Gli1+ mesenchymal stromal cells (MSCs) are recruited from the endosteal and perivascular niche to become fibrosis-driving myofibroblasts in the bone marrow. Genetic ablation of Gli1+ cells abolished BMF and rescued bone marrow failure. Pharmacological targeting of Gli proteins with GANT61 inhibited Gli1+ cell expansion and myofibroblast differentiation and attenuated fibrosis severity. The same pathway is also active in human BMF, and Gli1 expression in BMF significantly correlates with the severity of the disease. In addition, GANT61 treatment reduced the myofibroblastic phenotype of human MSCs isolated from patients with BMF, suggesting that targeting of Gli proteins could be a relevant therapeutic strategy.
Keywords: Gli1; bone marrow fibrosis; mesenchymal stem cells; myelofibrosis; myeloproliferative neoplasms.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures




Comment in
-
A new therapeutic target for myelofibrosis is cause for Gli.Sci Transl Med. 2017 May 31;9(392):eaan4297. doi: 10.1126/scitranslmed.aan4297. Sci Transl Med. 2017. PMID: 28566426 Free PMC article.
-
Gli-fully Halting the Progression of Fibrosis.Cell Stem Cell. 2017 Jun 1;20(6):735-736. doi: 10.1016/j.stem.2017.05.003. Cell Stem Cell. 2017. PMID: 28575684
References
-
- Abraham NG, Feldman E, Falck JR, Lutton JD, Schwartzman ML. Modulation of erythropoiesis by novel human bone marrow cytochrome P450-dependent metabolites of arachidonic acid. Blood. 1991;78:1461–1466. - PubMed
-
- Arranz L, Sanchez-Aguilera A, Martin-Perez D, Isern J, Langa X, Tzankov A, Lundberg P, Muntion S, Tzeng YS, Lai DM, et al. Neuropathy of haematopoietic stem cell niche is essential for myeloproliferative neoplasms. Nature. 2014;512:78–81. - PubMed
-
- Bares V, Ge X. gskb: Gene Set data for pathway analysis in mouse. 2015 R package version 161.
-
- Bhagwat N, Keller MD, Rampal RK, Shank K, de Stanchina E, Rose K, Amakye D, Levine RL. Improved Efficacy Of Comibnation Of JAK2 and Hedgehog Inhibitors in Myelofibrosis. Blood. 2013;122
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

