A genomic atlas of human adrenal and gonad development
- PMID: 28459107
- PMCID: PMC5407452
- DOI: 10.12688/wellcomeopenres.11253.2
A genomic atlas of human adrenal and gonad development
Abstract
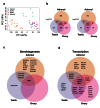
Background: In humans, the adrenal glands and gonads undergo distinct biological events between 6-10 weeks post conception (wpc), such as testis determination, the onset of steroidogenesis and primordial germ cell development. However, relatively little is currently known about the genetic mechanisms underlying these processes. We therefore aimed to generate a detailed genomic atlas of adrenal and gonad development across these critical stages of human embryonic and fetal development.
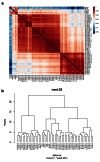
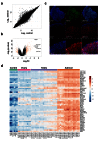
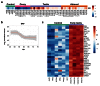
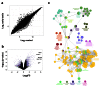
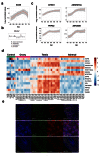
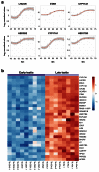
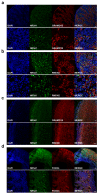
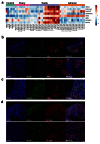
Methods: RNA was extracted from 53 tissue samples between 6-10 wpc (adrenal, testis, ovary and control). Affymetrix array analysis was performed and differential gene expression was analysed using Bioconductor. A mathematical model was constructed to investigate time-series changes across the dataset. Pathway analysis was performed using ClueGo and cellular localisation of novel factors confirmed using immunohistochemistry.
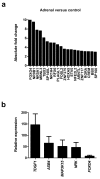
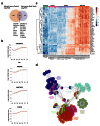
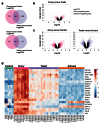
Results: Using this approach, we have identified novel components of adrenal development (e.g. ASB4, NPR3) and confirmed the role of SRY as the main human testis-determining gene. By mathematical modelling time-series data we have found new genes up-regulated with SOX9 in the testis (e.g. CITED1), which may represent components of the testis development pathway. We have shown that testicular steroidogenesis has a distinct onset at around 8 wpc and identified potential novel components in adrenal and testicular steroidogenesis (e.g. MGARP, FOXO4, MAP3K15, GRAMD1B, RMND2), as well as testis biomarkers (e.g. SCUBE1). We have also shown that the developing human ovary expresses distinct subsets of genes (e.g. OR10G9, OR4D5), but enrichment for established biological pathways is limited.
Conclusion: This genomic atlas is revealing important novel aspects of human development and new candidate genes for adrenal and reproductive disorders.
Keywords: adrenal; gene expression; germ cell; human development; ovary; sex development; steroidogenesis; testis.
Conflict of interest statement
Competing interests: No competing interests were disclosed.
Figures


References
-
- Achermann JC: A genomic atlas of human adrenal and gonad development. OSF. 2017. Data Source - DOI - PMC - PubMed
-
- Benjamini Y, Hochberg Y: Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57(1):289–300. Reference Source
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

