Enhanced nasopharyngeal infection and shedding associated with an epidemic lineage of emm3 group A Streptococcus
- PMID: 28459299
- PMCID: PMC5711448
- DOI: 10.1080/21505594.2017.1325070
Enhanced nasopharyngeal infection and shedding associated with an epidemic lineage of emm3 group A Streptococcus
Abstract
Background: A group A Streptococcus (GAS) lineage of genotype emm3, sequence type 15 (ST15) was associated with a 6 month upsurge in invasive GAS disease in the UK. The epidemic lineage (Lineage C) had lost 2 typical emm3 prophages, Φ315.1 and Φ315.2 associated with the superantigen ssa, but gained a different prophage (ΦUK-M3.1) associated with a different superantigen, speC and a DNAse spd1.
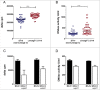
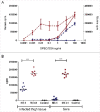
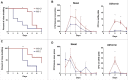
Methods and results: The presence of speC and spd1 in Lineage C ST15 strains enhanced both in vitro mitogenic and DNase activities over non-Lineage C ST15 strains. Invasive disease models in Galleria mellonella and SPEC-sensitive transgenic mice, revealed no difference in overall invasiveness of Lineage C ST15 strains compared with non-Lineage C ST15 strains, consistent with clinical and epidemiological analysis. Lineage C strains did however markedly prolong murine nasal infection with enhanced nasal and airborne shedding compared with non-Lineage C strains. Deletion of speC or spd1 in 2 Lineage C strains identified a possible role for spd1 in airborne shedding from the murine nasopharynx.
Conclusions: Nasopharyngeal infection and shedding of Lineage C strains was enhanced compared with non-Lineage C strains and this was, in part, mediated by the gain of the DNase spd1 through prophage acquisition.
Keywords: DNases; Streptococcus pyogenes; epidemic upsurge; genotype emm3; group A Streptococcus; invasive group A streptococcal disease; nasopharyngeal infection; prophage; serotype M3; superantigens.
Figures



Comment in
-
Unravelling pathogenetic mechanisms of epidemic lineages.Virulence. 2017 Oct 3;8(7):1102-1104. doi: 10.1080/21505594.2017.1328344. Epub 2017 May 10. Virulence. 2017. PMID: 28489957 Free PMC article. No abstract available.
References
-
- Davies MR, Holden MT, Coupland P, Chen JH, Venturini C, Barnett TC, Zakour NL, Tse H, Dougan G, Yuen KY, et al.. Emergence of scarlet fever Streptococcus pyogenes emm12 clones in Hong Kong is associated with toxin acquisition and multidrug resistance. Nat Genet 2015; 47:84-7; PMID:25401300; https://doi.org/ 10.1038/ng.3147 - DOI - PubMed
-
- Beres SB, Carroll RK, Shea PR, Sitkiewicz I, Martinez-Gutierrez JC, Low DE, McGeer A, Willey BM, Green K, Tyrrell GJ, et al.. Molecular complexity of successive bacterial epidemics deconvoluted by comparative pathogenomics. Proc Natl Acad Sci USA 2010; 107:4371-6; PMID:20142485; https://doi.org/ 10.1073/pnas.0911295107 - DOI - PMC - PubMed
-
- Health Protection Agency Enhanced surveillance intitiated for group A streptococcal infections. Health Protection Report 2009a; 3(8) Available at: http://webarchive.nationalarchives.gov.uk/20140714084352/http://www.hpa.org.uk/hpr/archives/2009/news0809.htm#gas0809
-
- Health Protection Agency Group A streptococcal infections: fourth update on seasonal activity, 2008/09. Health Protection Report 2009b; 3(29) Available at: http://webarchive.nationalarchives.gov.uk/20140714084352/http://www.hpa....
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
