Structure of RNA Stem Loop B from the Picornavirus Replication Platform
- PMID: 28459542
- PMCID: PMC5699211
- DOI: 10.1021/acs.biochem.7b00141
Structure of RNA Stem Loop B from the Picornavirus Replication Platform
Abstract
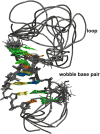
The presumptive RNA cloverleaf at the start of the 5'-untranslated region of the picornavirus genome is an essential element in replication. Stem loop B (SLB) of the cloverleaf is a recognition site for the host polyC-binding protein, which initiates a switch from translation to replication. Here we present the solution structure of human rhinovirus isotype 14 SLB using nuclear magnetic resonance spectroscopy. SLB adopts a predominantly A-form helical structure. The stem contains five Watson-Crick base pairs and one wobble base pair and is capped by an eight-nucleotide loop. The wobble base pair introduces perturbations into the helical parameters but does not appear to introduce flexibility. However, the helix major groove appears to be accessible. Flexibility is seen throughout the loop and in the terminal nucleotides. The pyrimidine-rich region of the loop, the apparent recognition site for the polyC-binding protein, is the most disordered region of the structure.
Conflict of interest statement
Figures





References
-
- Knowles N, Hovi T, King A, Stanway G. The picornaviruses. American Society for Microbiology Press; Washington, DC: 2010. Overview of taxonomy; pp. 19–32.
-
- Greenberg SB. Respiratory consequences of rhinovirus infection. Arch Intern Med. 2003;163:278–284. - PubMed
-
- Agol VI, Paul AV, Wimmer E. Paradoxes of the replication of picornaviral genomes. Virus Res. 1999;62:129–147. - PubMed
-
- Andino R, Rieckhof GE, Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

