Global miRNA expression analysis identifies novel key regulators of plasma cell differentiation and malignant plasma cell
- PMID: 28459970
- PMCID: PMC5449613
- DOI: 10.1093/nar/gkx327
Global miRNA expression analysis identifies novel key regulators of plasma cell differentiation and malignant plasma cell
Abstract
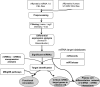
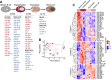
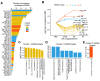
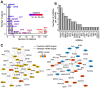
MicroRNAs (miRNAs) are small noncoding RNAs that attenuate expression of their mRNA targets. Here, we developed a new method and an R package, to easily infer candidate miRNA-mRNA target interactions that could be functional during a given biological process. Using this method, we described, for the first time, a comprehensive integrated analysis of miRNAs and mRNAs during human normal plasma cell differentiation (PCD). Our results reveal 63 miRNAs with significant temporal changes in their expression during normal PCD. We derived a high-confidence network of 295 target relationships comprising 47 miRNAs and 141 targets. These relationships include new examples of miRNAs that appear to coordinately regulate multiple members of critical pathways associated with PCD. Consistent with this, we have experimentally validated a role for the miRNA-30b/c/d-mediated regulation of key PCD factors (IRF4, PRDM1, ELL2 and ARID3A). Furthermore, we found that 24 PCD stage-specific miRNAs are aberrantly overexpressed in multiple myeloma (MM) tumor plasma cells compared to their normal counterpart, suggesting that MM cells frequently acquired expression changes in miRNAs already undergoing dynamic expression modulation during normal PCD. Altogether, our analysis identifies candidate novel key miRNAs regulating networks of significance for normal PCD and malignant plasma cell biology.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Shapiro-Shelef M., Calame K.. Regulation of plasma-cell development. Nat. Rev. Immunol. 2005; 5:230–242. - PubMed
-
- Jourdan M., Caraux A., Caron G., Robert N., Fiol G., Reme T., Bollore K., Vendrell J.P., Le Gallou S., Mourcin F. et al. . Characterization of a transitional preplasmablast population in the process of human B cell to plasma cell differentiation. J. Immunol. 2011; 187:3931–3941. - PubMed
-
- Jourdan M., Cren M., Robert N., Bollore K., Fest T., Duperray C., Guilloton F., Hose D., Tarte K., Klein B.. IL-6 supports the generation of human long-lived plasma cells in combination with either APRIL or stromal cell-soluble factors. Leukemia. 2014; 28:1647–1656. - PubMed
-
- Nutt S.L., Hodgkin P.D., Tarlinton D.M., Corcoran L.M.. The generation of antibody-secreting plasma cells. Nat. Rev. Immunol. 2015; 15:160–171. - PubMed
-
- Shaffer A.L., Lin K.I., Kuo T.C., Yu X., Hurt E.M., Rosenwald A., Giltnane J.M., Yang L., Zhao H., Calame K. et al. . Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002; 17:51–62. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

