Interactive microbial distribution analysis using BioAtlas
- PMID: 28460071
- PMCID: PMC5570126
- DOI: 10.1093/nar/gkx304
Interactive microbial distribution analysis using BioAtlas
Abstract
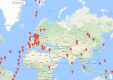
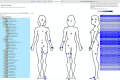
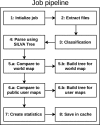
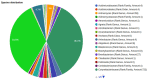
Massive amounts of 16S rRNA sequencing data have been stored in publicly accessible databases, such as GOLD, SILVA, GreenGenes (GG), and the Ribosomal Database Project (RDP). Many of these sequences are tagged with geo-locations. Nevertheless, researchers currently lack a user-friendly tool to analyze microbial distribution in a location-specific context. BioAtlas is an interactive web application that closes this gap between sequence databases, taxonomy profiling and geo/body-location information. It enables users to browse taxonomically annotated sequences across (i) the world map, (ii) human body maps and (iii) user-defined maps. It further allows for (iv) uploading of own sample data, which can be placed on existing maps to (v) browse the distribution of the associated taxonomies. Finally, BioAtlas enables users to (vi) contribute custom maps (e.g. for plants or animals) and to map taxonomies to pre-defined map locations. In summary, BioAtlas facilitates map-supported browsing of public 16S rRNA sequence data and analyses of user-provided sequences without requiring manual mapping to taxonomies and existing databases.
Availability: http://bioatlas.compbio.sdu.dk/.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Green J., Bohannan B.J.. Spatial scaling of microbial biodiversity. Trends Ecol. Evol. 2006; 21:501–507. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

