Decreased ATF4 expression as a mechanism of acquired resistance to long-term amino acid limitation in cancer cells
- PMID: 28460466
- PMCID: PMC5432347
- DOI: 10.18632/oncotarget.15828
Decreased ATF4 expression as a mechanism of acquired resistance to long-term amino acid limitation in cancer cells
Abstract
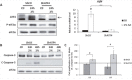
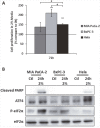
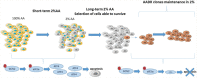
The uncontrolled growth of tumor can lead to the formation of area deprived in nutrients. Due to their high genetic instability, tumor cells can adapt and develop resistance to this pro-apoptotic environment. Among the resistance mechanisms, those involved in the resistance to long-term amino acid restriction are not elucidated. A long-term amino acid restriction is particularly deleterious since nine of them cannot be synthetized by the cells. In order to determine how cancer cells face a long-term amino acid deprivation, we developed a cell model selected for its capacity to resist a long-term amino acid limitation. We exerted a selection pressure on mouse embryonic fibroblast to isolate clones able to survive with low amino acid concentration. The study of several clones revealed an alteration of the eiF2α/ATF4 pathway. Compared to the parental cells, the clones exhibited a decreased expression of the transcription factor ATF4 and its target genes. Likewise, the knock-down of ATF4 in parental cells renders them resistant to amino acid deprivation. Moreover, this association between a low level of ATF4 protein and the resistance to amino acid deprivation was also observed in the cancer cell line BxPC-3. This resistance was abolished when ATF4 was overexpressed. Therefore, decreasing ATF4 expression may be one important mechanism for cancer cells to survive under prolonged amino acid deprivation.
Keywords: ATF4; amino acids; apoptosis; cancer; resistance.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures




References
-
- Vaupel P, Kallinowski F, Okunieff P. Blood Flow, Oxygen and Nutrient Supply, and Metabolic Microenvironment of Human Tumors: A Review. Cancer Res. 1989;49:6449–65. - PubMed
-
- Graeber TG, Osmanian C, Jacks T, Housman DE, Koch CJ, Lowe SW, Giaccia AJ. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature. 1996;379:88–91. - PubMed
-
- Bristow RG, Hill RP. Hypoxia and metabolism: Hypoxia, DNA repair and genetic instability. Nat Rev Cancer. 2008;8:180–92. - PubMed
-
- Pouysségur J, Dayan F, Mazure NM. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature. 2006;441:437–43. - PubMed
-
- Harris AL. Hypoxia — a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

