Transcriptomic profiling and quantitative high-throughput (qHTS) drug screening of CDH1 deficient hereditary diffuse gastric cancer (HDGC) cells identify treatment leads for familial gastric cancer
- PMID: 28460635
- PMCID: PMC5412046
- DOI: 10.1186/s12967-017-1197-5
Transcriptomic profiling and quantitative high-throughput (qHTS) drug screening of CDH1 deficient hereditary diffuse gastric cancer (HDGC) cells identify treatment leads for familial gastric cancer
Abstract
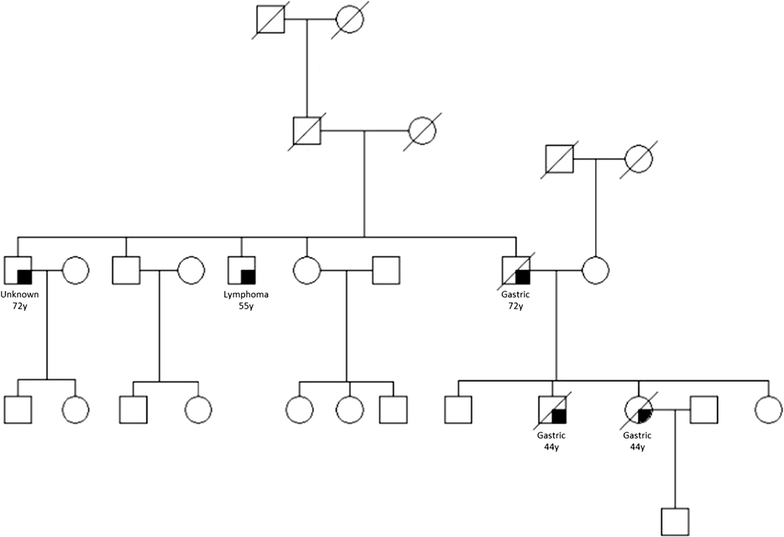
Background: Patients with hereditary diffuse gastric cancer (HDGC), a cancer predisposition syndrome associated with germline mutations of the CDH1 (E-cadherin) gene, have few effective treatment options. Despite marked differences in natural history, histopathology, and genetic profile to patients afflicted by sporadic gastric cancer, patients with HDGC receive, in large, identical systemic regimens. The lack of a robust preclinical in vitro system suitable for effective drug screening has been one of the obstacles to date which has hampered therapeutic advances in this rare disease.
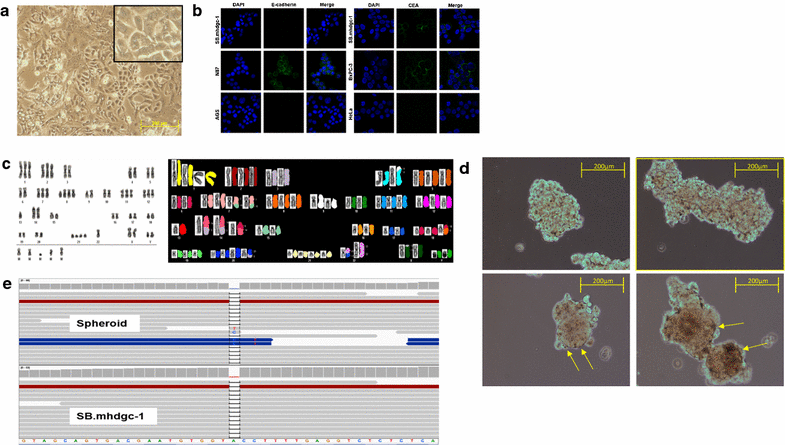
Methods: In order to identify therapeutic leads selective for the HDGC subtype of gastric cancer, we compared gene expression profiles and drug phenotype derived from an oncology library of 1912 compounds between gastric cancer cells established from a patient with metastatic HDGC harboring a c.1380delA CDH1 germline variant and sporadic gastric cancer cells.
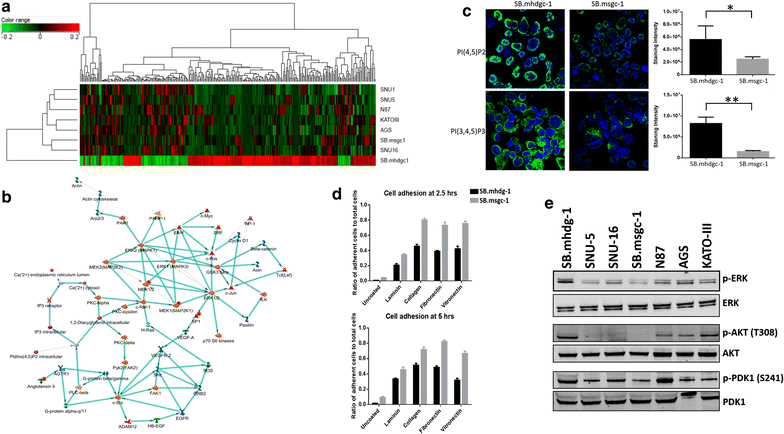
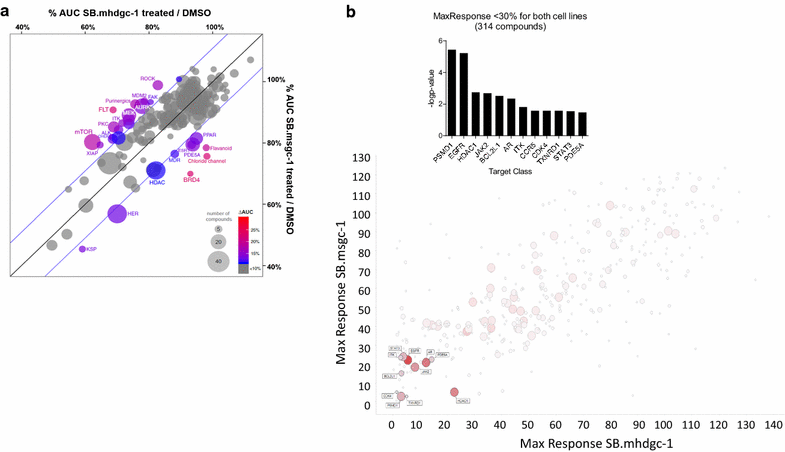
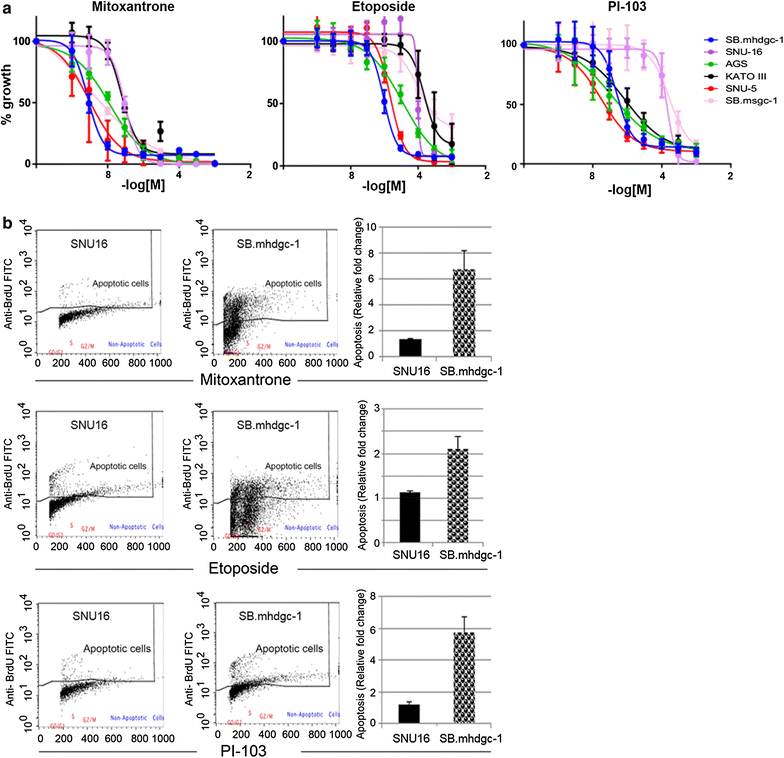
Results: Unsupervised hierarchical cluster analysis shows select gene expression alterations in c.1380delA CDH1 SB.mhdgc-1 cells compared to a panel of sporadic gastric cancer cell lines with enrichment of ERK1-ERK2 (extracellular signal regulated kinase) and IP3 (inositol trisphosphate)/DAG (diacylglycerol) signaling as the top networks in c.1380delA SB.mhdgc-1 cells. Intracellular phosphatidylinositol intermediaries were increased upon direct measure in c.1380delA CDH1 SB.mhdgc-1 cells. Differential high-throughput drug screening of c.1380delA CDH1 SB.mhdgc-1 versus sporadic gastric cancer cells identified several compound classes with enriched activity in c.1380 CDH1 SB.mhdgc-1 cells including mTOR (Mammalian Target Of Rapamycin), MEK (Mitogen-Activated Protein Kinase), c-Src kinase, FAK (Focal Adhesion Kinase), PKC (Protein Kinase C), or TOPO2 (Topoisomerase II) inhibitors. Upon additional drug response testing, dual PI3K (Phosphatidylinositol 3-Kinase)/mTOR and topoisomerase 2A inhibitors displayed up to >100-fold increased activity in hereditary c.1380delA CDH1 gastric cancer cells inducing apoptosis most effectively in cells with deficient CDH1 function.
Conclusion: Integrated pharmacological and transcriptomic profiling of hereditary diffuse gastric cancer cells with a loss-of-function c.1380delA CDH1 mutation implies various pharmacological vulnerabilities selective to CDH1-deficient familial gastric cancer cells and suggests novel treatment leads for future preclinical and clinical treatment studies of familial gastric cancer.
Keywords: Differential gene expression profiling; Hereditary diffuse gastric cancer (HDGC); High throughput drug screening; Therapeutic leads; c.1380delA CDH1 gastric cancer cells.
Figures
Similar articles
-
Merging perspectives: genotype-directed molecular therapy for hereditary diffuse gastric cancer (HDGC) and E-cadherin-EGFR crosstalk.Clin Transl Med. 2018 Feb 22;7(1):7. doi: 10.1186/s40169-018-0184-7. Clin Transl Med. 2018. PMID: 29468433 Free PMC article.
-
Accuracy of Hereditary Diffuse Gastric Cancer Testing Criteria and Outcomes in Patients With a Germline Mutation in CDH1.Gastroenterology. 2015 Oct;149(4):897-906.e19. doi: 10.1053/j.gastro.2015.06.003. Epub 2015 Jun 11. Gastroenterology. 2015. PMID: 26072394
-
Cleft lip/palate and hereditary diffuse gastric cancer: report of a family harboring a CDH1 c.687 + 1G > A germline mutation and review of the literature.Fam Cancer. 2019 Apr;18(2):253-260. doi: 10.1007/s10689-018-0111-5. Fam Cancer. 2019. PMID: 30306390 Review.
-
Hereditary Diffuse Gastric Cancer Syndrome: CDH1 Mutations and Beyond.JAMA Oncol. 2015 Apr;1(1):23-32. doi: 10.1001/jamaoncol.2014.168. JAMA Oncol. 2015. PMID: 26182300
-
CDH1 and hereditary diffuse gastric cancer: a narrative review.Chin Clin Oncol. 2023 Jun;12(3):25. doi: 10.21037/cco-23-36. Epub 2023 Jun 5. Chin Clin Oncol. 2023. PMID: 37303221 Review.
Cited by
-
Family's History Based on the CDH1 Germline Variant (c.360delG) and a Suspected Hereditary Gastric Cancer Form.Int J Mol Sci. 2020 Jul 11;21(14):4904. doi: 10.3390/ijms21144904. Int J Mol Sci. 2020. PMID: 32664545 Free PMC article.
-
Merging perspectives: genotype-directed molecular therapy for hereditary diffuse gastric cancer (HDGC) and E-cadherin-EGFR crosstalk.Clin Transl Med. 2018 Feb 22;7(1):7. doi: 10.1186/s40169-018-0184-7. Clin Transl Med. 2018. PMID: 29468433 Free PMC article.
-
Higher risk of gastric cancer among immigrants to Ontario: a population-based matched cohort study with over 2 million individuals.Gastric Cancer. 2018 Jul;21(4):588-597. doi: 10.1007/s10120-017-0790-x. Epub 2017 Dec 28. Gastric Cancer. 2018. PMID: 29285629
-
Quality Control of Quantitative High Throughput Screening Data.Front Genet. 2019 May 9;10:387. doi: 10.3389/fgene.2019.00387. eCollection 2019. Front Genet. 2019. PMID: 31143201 Free PMC article.
-
Current advances in understanding the molecular profile of hereditary diffuse gastric cancer and its clinical implications.J Exp Clin Cancer Res. 2023 Mar 4;42(1):57. doi: 10.1186/s13046-023-02622-3. J Exp Clin Cancer Res. 2023. PMID: 36869400 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

