Mass Spectrometric Analyses Reveal a Central Role for Ubiquitylation in Remodeling the Arabidopsis Proteome during Photomorphogenesis
- PMID: 28461270
- PMCID: PMC5695678
- DOI: 10.1016/j.molp.2017.04.008
Mass Spectrometric Analyses Reveal a Central Role for Ubiquitylation in Remodeling the Arabidopsis Proteome during Photomorphogenesis
Abstract
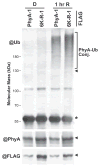
The switch from skotomorphogenesis to photomorphogenesis is a key developmental transition in the life of seed plants. While much of the underpinning proteome remodeling is driven by light-induced changes in gene expression, the proteolytic removal of specific proteins by the ubiquitin-26S proteasome system is also likely paramount. Through mass spectrometric analysis of ubiquitylated proteins affinity-purified from etiolated Arabidopsis seedlings before and after red-light irradiation, we identified a number of influential proteins whose ubiquitylation status is modified during this switch. We observed a substantial enrichment for proteins involved in auxin, abscisic acid, ethylene, and brassinosteroid signaling, peroxisome function, disease resistance, protein phosphorylation and light perception, including the phytochrome (Phy) A and phototropin photoreceptors. Soon after red-light treatment, PhyA becomes the dominant ubiquitylated species, with ubiquitin attachment sites mapped to six lysines. A PhyA mutant protected from ubiquitin addition at these sites is substantially more stable in planta upon photoconversion to Pfr and is hyperactive in driving photomorphogenesis. However, light still stimulates ubiquitylation and degradation of this mutant, implying that other attachment sites and/or proteolytic pathways exist. Collectively, we expand the catalog of ubiquitylation targets in Arabidopsis and show that this post-translational modification is central to the rewiring of plants for photoautotrophic growth.
Keywords: Arabidopsis; mass spectrometry; photomorphogenesis; phytochrome degradation; ubiquitin.
Copyright © 2017 The Author. Published by Elsevier Inc. All rights reserved.
Figures








References
-
- Abramoff M, Magalhaes P, Ram S. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42.
-
- Al-Sady B, Ni W, Kircher S, Schafer E, Quail PH. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell. 2006;23:439–446. - PubMed
-
- Bernhardt A, Lechner E, Hano P, Schade V, Dieterle M, Anders M, Dubin MJ, Benvenuto G, Bowler C, Genschik P, et al. CUL4 associates with DDB1 and DET1 and its downregulation affects diverse aspects of development in Arabidopsis thaliana. Plant J. 2006;47:591–603. - PubMed
-
- Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

