Retrospective analysis of natural products provides insights for future discovery trends
- PMID: 28461474
- PMCID: PMC5465889
- DOI: 10.1073/pnas.1614680114
Retrospective analysis of natural products provides insights for future discovery trends
Abstract
Understanding of the capacity of the natural world to produce secondary metabolites is important to a broad range of fields, including drug discovery, ecology, biosynthesis, and chemical biology, among others. Both the absolute number and the rate of discovery of natural products have increased significantly in recent years. However, there is a perception and concern that the fundamental novelty of these discoveries is decreasing relative to previously known natural products. This study presents a quantitative examination of the field from the perspective of both number of compounds and compound novelty using a dataset of all published microbial and marine-derived natural products. This analysis aimed to explore a number of key questions, such as how the rate of discovery of new natural products has changed over the past decades, how the average natural product structural novelty has changed as a function of time, whether exploring novel taxonomic space affords an advantage in terms of novel compound discovery, and whether it is possible to estimate how close we are to having described all of the chemical space covered by natural products. Our analyses demonstrate that most natural products being published today bear structural similarity to previously published compounds, and that the range of scaffolds readily accessible from nature is limited. However, the analysis also shows that the field continues to discover appreciable numbers of natural products with no structural precedent. Together, these results suggest that the development of innovative discovery methods will continue to yield compounds with unique structural and biological properties.
Keywords: chemical diversity; chemoinformatics; drug discovery; natural products; structural similarity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


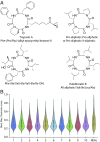


Comment in
-
The natural productome.Proc Natl Acad Sci U S A. 2017 May 30;114(22):5564-5566. doi: 10.1073/pnas.1706266114. Epub 2017 May 22. Proc Natl Acad Sci U S A. 2017. PMID: 28533417 Free PMC article. No abstract available.
-
Reply to Skinnider and Magarvey: Rates of novel natural product discovery remain high.Proc Natl Acad Sci U S A. 2017 Aug 1;114(31):E6273. doi: 10.1073/pnas.1711139114. Epub 2017 Jul 14. Proc Natl Acad Sci U S A. 2017. PMID: 28710331 Free PMC article. No abstract available.
-
Statistical reanalysis of natural products reveals increasing chemical diversity.Proc Natl Acad Sci U S A. 2017 Aug 1;114(31):E6271-E6272. doi: 10.1073/pnas.1708560114. Epub 2017 Jul 14. Proc Natl Acad Sci U S A. 2017. PMID: 28710332 Free PMC article. No abstract available.
References
-
- Newman DJ, Cragg GM. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod. 2016;79:629–661. - PubMed
-
- Harvey AL, Edrada-Ebel R, Quinn RJ. The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discov. 2015;14:111–129. - PubMed
-
- Gwynn MN, Portnoy A, Rittenhouse SF, Payne DJ. Challenges of antibacterial discovery revisited. Ann N Y Acad Sci. 2010;1213:5–19. - PubMed
-
- Kong D-X, Guo M-Y, Xiao Z-H, Chen L-L, Zhang H-Y. Historical variation of structural novelty in a natural product library. Chem Biodivers. 2011;8:1968–1977. - PubMed
-
- Walsh CT. A chemocentric view of the natural product inventory. Nat Chem Biol. 2015;11:620–624. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

