Pre/pro-B cells generate macrophage populations during homeostasis and inflammation
- PMID: 28461481
- PMCID: PMC5441795
- DOI: 10.1073/pnas.1616417114
Pre/pro-B cells generate macrophage populations during homeostasis and inflammation
Abstract
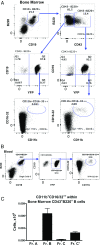
Most tissue-resident macrophages (Mφs) are believed to be derived prenatally and are assumed to maintain themselves throughout life by self-proliferation. However, in adult mice we identified a progenitor within bone marrow, early pro-B cell/fraction B, that differentiates into tissue Mφs. These Mφ precursors have non-rearranged B-cell receptor genes and coexpress myeloid (GR1, CD11b, and CD16/32) and lymphoid (B220 and CD19) lineage markers. During steady state, these precursors exit bone marrow, losing Gr1, and enter the systemic circulation, seeding the gastrointestinal system as well as pleural and peritoneal cavities but not the brain. While in these tissues, they acquire a transcriptome identical to embryonically derived tissue-resident Mφs. Similarly, these Mφ precursors also enter sites of inflammation, gaining CD115, F4/80, and CD16/32, and become indistinguishable from blood monocyte-derived Mφs. Thus, we have identified a population of cells within the bone marrow early pro-B cell compartment that possess functional plasticity to differentiate into either tissue-resident or inflammatory Mφs, depending on microenvironmental signals. We propose that these precursors represent an additional source of Mφ populations in adult mice during steady state and inflammation.
Keywords: homeostasis; inflammation; macrophages.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
Publication types
MeSH terms
Grants and funding
- WT_/Wellcome Trust/United Kingdom
- BBS/E/D/05191131/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/20211552/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/20211553/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

