Protein-mediated viral latency is a novel mechanism for Merkel cell polyomavirus persistence
- PMID: 28461484
- PMCID: PMC5441811
- DOI: 10.1073/pnas.1703879114
Protein-mediated viral latency is a novel mechanism for Merkel cell polyomavirus persistence
Abstract
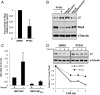
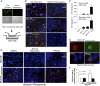
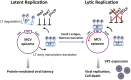
Viral latency, in which a virus genome does not replicate independently of the host cell genome and produces no infectious particles, is required for long-term virus persistence. There is no known latency mechanism for chronic small DNA virus infections. Merkel cell polyomavirus (MCV) causes an aggressive skin cancer after prolonged infection and requires an active large T (LT) phosphoprotein helicase to replicate. We show that evolutionarily conserved MCV LT phosphorylation sites are constitutively recognized by cellular Fbw7, βTrCP, and Skp2 Skp-F-box-cullin (SCF) E3 ubiquitin ligases, which degrade and suppress steady-state LT protein levels. Knockdown of each of these E3 ligases enhances LT stability and promotes MCV genome replication. Mutations at two of these phosphoreceptor sites [serine (S)220 and S239] in the full viral genome increase LT levels and promote MCV virion production and transmission, which can be neutralized with anti-capsid antibody. Virus activation is not mediated by viral gene transactivation, given that these mutations do not increase late gene transcription in the absence of genome replication. Mechanistic target of rapamycin inhibition by either nutrient starvation or use of an active site inhibitor reduces Skp2 levels and stabilizes LT, leading to enhanced MCV replication and transmission. MCV can sense stresses in its intracellular environment, such as nutrient loss, through SCF E3 ligase activities, and responds by initiating active viral transmission. Protein-mediated viral latency through cellular SCF E3 ligase targeting of viral replication proteins is a unique form of latency that may promote chronic viral persistence for some small DNA and RNA viruses.
Keywords: E3 ligase; Merkel cell polyomavirus; large T; latency; transmission.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

