Brd4 modulates the innate immune response through Mnk2-eIF4E pathway-dependent translational control of IκBα
- PMID: 28461486
- PMCID: PMC5441817
- DOI: 10.1073/pnas.1700109114
Brd4 modulates the innate immune response through Mnk2-eIF4E pathway-dependent translational control of IκBα
Abstract
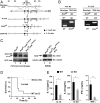
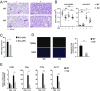
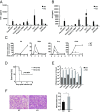
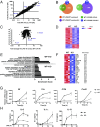
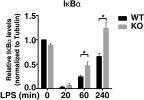
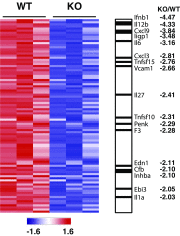
Bromodomain-containing factor Brd4 has emerged as an important transcriptional regulator of NF-κB-dependent inflammatory gene expression. However, the in vivo physiological function of Brd4 in the inflammatory response remains poorly defined. We now demonstrate that mice deficient for Brd4 in myeloid-lineage cells are resistant to LPS-induced sepsis but are more susceptible to bacterial infection. Gene-expression microarray analysis of bone marrow-derived macrophages (BMDMs) reveals that deletion of Brd4 decreases the expression of a significant amount of LPS-induced inflammatory genes while reversing the expression of a small subset of LPS-suppressed genes, including MAP kinase-interacting serine/threonine-protein kinase 2 (Mknk2). Brd4-deficient BMDMs display enhanced Mnk2 expression and the corresponding eukaryotic translation initiation factor 4E (eIF4E) activation after LPS stimulation, leading to an increased translation of IκBα mRNA in polysomes. The enhanced newly synthesized IκBα reduced the binding of NF-κB to the promoters of inflammatory genes, resulting in reduced inflammatory gene expression and cytokine production. By modulating the translation of IκBα via the Mnk2-eIF4E pathway, Brd4 provides an additional layer of control for NF-κB-dependent inflammatory gene expression and inflammatory response.
Keywords: Brd4; IκBα resynthesis; Mnk2; NF-κB; eIF4E.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: Evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. - PubMed
-
- Baldwin AS., Jr The NF-kappa B and I kappa B proteins: New discoveries and insights. Annu Rev Immunol. 1996;14:649–683. - PubMed
-
- Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. - PubMed
-
- Chen LF, Greene WC. Shaping the nuclear action of NF-kappaB. Nat Rev Mol Cell Biol. 2004;5:392–401. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

