Nuclear export of misfolded SOD1 mediated by a normally buried NES-like sequence reduces proteotoxicity in the nucleus
- PMID: 28463106
- PMCID: PMC5449186
- DOI: 10.7554/eLife.23759
Nuclear export of misfolded SOD1 mediated by a normally buried NES-like sequence reduces proteotoxicity in the nucleus
Abstract
Over 170 different mutations in the gene encoding SOD1 all cause amyotrophic lateral sclerosis (ALS). Available studies have been primarily focused on the mechanisms underlying mutant SOD1 cytotoxicity. How cells defend against the cytotoxicity remains largely unknown. Here, we show that misfolding of ALS-linked SOD1 mutants and wild-type (wt) SOD1 exposes a normally buried nuclear export signal (NES)-like sequence. The nuclear export carrier protein CRM1 recognizes this NES-like sequence and exports misfolded SOD1 to the cytoplasm. Antibodies against the NES-like sequence recognize misfolded SOD1, but not native wt SOD1 both in vitro and in vivo. Disruption of the NES consensus sequence relocalizes mutant SOD1 to the nucleus, resulting in higher toxicity in cells, and severer impairments in locomotion, egg-laying, and survival in Caenorhabditis elegans. Our data suggest that SOD1 mutants are removed from the nucleus by CRM1 as a defense mechanism against proteotoxicity of misfolded SOD1 in the nucleus.
Keywords: ALS; C. elegans; SOD1; cell biology; cytotoxicity; nuclear export signal; protein misfolding.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures
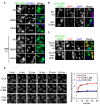
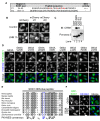
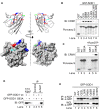

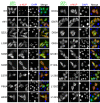
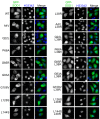
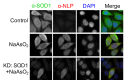
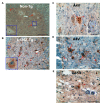
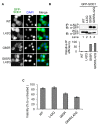


Similar articles
-
MIF inhibits the formation and toxicity of misfolded SOD1 amyloid aggregates: implications for familial ALS.Cell Death Dis. 2018 Jan 25;9(2):107. doi: 10.1038/s41419-017-0130-4. Cell Death Dis. 2018. PMID: 29371591 Free PMC article.
-
Generation of influenza A virus NS2 (NEP) mutants with an altered nuclear export signal sequence.J Virol. 2004 Sep;78(18):10149-55. doi: 10.1128/JVI.78.18.10149-10155.2004. J Virol. 2004. PMID: 15331747 Free PMC article.
-
Using a Simple Cellular Assay to Map NES Motifs in Cancer-Related Proteins, Gain Insight into CRM1-Mediated NES Export, and Search for NES-Harboring Micropeptides.Int J Mol Sci. 2020 Sep 1;21(17):6341. doi: 10.3390/ijms21176341. Int J Mol Sci. 2020. PMID: 32882917 Free PMC article.
-
SOD1 in neurotoxicity and its controversial roles in SOD1 mutation-negative ALS.Adv Biol Regul. 2016 Jan;60:95-104. doi: 10.1016/j.jbior.2015.10.006. Epub 2015 Oct 31. Adv Biol Regul. 2016. PMID: 26563614 Review.
-
Oxidized/misfolded superoxide dismutase-1: the cause of all amyotrophic lateral sclerosis?Ann Neurol. 2007 Dec;62(6):553-9. doi: 10.1002/ana.21319. Ann Neurol. 2007. PMID: 18074357 Review.
Cited by
-
An update to the CRM1 cargo/NES database NESdb.Mol Biol Cell. 2021 Mar 15;32(6):467-469. doi: 10.1091/mbc.E20-11-0694. Mol Biol Cell. 2021. PMID: 33720780 Free PMC article. No abstract available.
-
Nuclear pore dysfunction and disease: a complex opportunity.Nucleus. 2024 Dec;15(1):2314297. doi: 10.1080/19491034.2024.2314297. Epub 2024 Feb 21. Nucleus. 2024. PMID: 38383349 Free PMC article. Review.
-
RNA Molecular Signature Profiling in PBMCs of Sporadic ALS Patients: HSP70 Overexpression Is Associated with Nuclear SOD1.Cells. 2022 Jan 15;11(2):293. doi: 10.3390/cells11020293. Cells. 2022. PMID: 35053410 Free PMC article.
-
Nicotinamide Riboside Enhances Mitochondrial Proteostasis and Adult Neurogenesis through Activation of Mitochondrial Unfolded Protein Response Signaling in the Brain of ALS SOD1G93A Mice.Int J Biol Sci. 2020 Jan 1;16(2):284-297. doi: 10.7150/ijbs.38487. eCollection 2020. Int J Biol Sci. 2020. PMID: 31929756 Free PMC article.
-
Structural Properties and Interaction Partners of Familial ALS-Associated SOD1 Mutants.Front Neurol. 2019 May 21;10:527. doi: 10.3389/fneur.2019.00527. eCollection 2019. Front Neurol. 2019. PMID: 31164862 Free PMC article. Review.
References
-
- Bali T, Self W, Liu J, Siddique T, Wang LH, Bird TD, Ratti E, Atassi N, Boylan KB, Glass JD, Maragakis NJ, Caress JB, McCluskey LF, Appel SH, Wymer JP, Gibson S, Zinman L, Mozaffar T, Callaghan B, McVey AL, Jockel-Balsarotti J, Allred P, Fisher ER, Lopate G, Pestronk A, Cudkowicz ME, Miller TM. Defining SOD1 ALS natural history to guide therapeutic clinical trial design. Journal of Neurology, Neurosurgery & Psychiatry. 2017;88:99–105. doi: 10.1136/jnnp-2016-313521. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

