Comparative interactomics provides evidence for functional specialization of the nuclear pore complex
- PMID: 28463551
- PMCID: PMC5597298
- DOI: 10.1080/19491034.2017.1313936
Comparative interactomics provides evidence for functional specialization of the nuclear pore complex
Abstract
The core architecture of the eukaryotic cell was established well over one billion years ago, and is largely retained in all extant lineages. However, eukaryotic cells also possess lineage-specific features, frequently keyed to specific functional requirements. One quintessential core eukaryotic structure is the nuclear pore complex (NPC), responsible for regulating exchange of macromolecules between the nucleus and cytoplasm as well as acting as a nuclear organizational hub. NPC architecture has been best documented in one eukaryotic supergroup, the Opisthokonts (e.g. Saccharomyces cerevisiae and Homo sapiens), which although compositionally similar, have significant variations in certain NPC subcomplex structures. The variation of NPC structure across other taxa in the eukaryotic kingdom however, remains poorly understood. We explored trypanosomes, highly divergent organisms, and mapped and assigned their NPC proteins to specific substructures to reveal their NPC architecture. We showed that the NPC central structural scaffold is conserved, likely across all eukaryotes, but more peripheral elements can exhibit very significant lineage-specific losses, duplications or other alterations in their components. Amazingly, trypanosomes lack the major components of the mRNA export platform that are asymmetrically localized within yeast and vertebrate NPCs. Concomitant with this, the trypanosome NPC is ALMOST completely symmetric with the nuclear basket being the only major source of asymmetry. We suggest these features point toward a stepwise evolution of the NPC in which a coating scaffold first stabilized the pore after which selective gating emerged and expanded, leading to the addition of peripheral remodeling machineries on the nucleoplasmic and cytoplasmic sides of the pore.
Keywords: Trypanosoma brucei; eukaryogenesis; mRNA export; molecular evolution; nuclear pore complex.
Figures
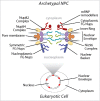
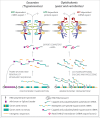
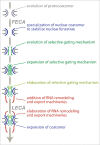
Comment on
- Extra view to: Obado SO, Brillantes M, Uryu K, Zhang W, Ketaren NE, Chait BT, Field MC, Rout MP. Interactome mapping reveals the evolutionary history of the nuclear pore complex. PLoS Biol. 2016; 14(2):e1002365; PMID:; https://doi.org/10.1371/journal.pbio.1002365. doi: 10.1371/journal.pbio.1002365
References
-
- Cavalier-Smith T. Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biology letters 2010; 6:342-5; PMID:20031978; https://doi.org/10.1098/rsbl.2009.0948 - DOI - PMC - PubMed
-
- Adl SM, Simpson AG, Lane CE, Lukes J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, et al.. The revised classification of eukaryotes. J Eukaryot Microbiol 2012; 59:429-93; PMID:23020233; https://doi.org/10.1111/j.1550-7408.2012.00644.x - DOI - PMC - PubMed
-
- Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol 2002; 158:915-27; PMID:12196509; https://doi.org/10.1083/jcb.200206106 - DOI - PMC - PubMed
-
- DeGrasse JA, DuBois KN, Devos D, Siegel TN, Sali A, Field MC, Rout MP, Chait BT. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol Cell Proteomics : MCP 2009; 8:2119-30; https://doi.org/10.1074/mcp.M900038-MCP200 - DOI - PMC - PubMed
-
- Obado SO, Brillantes M, Uryu K, Zhang W, Ketaren NE, Chait BT, Field MC, Rout MP. Interactome Mapping Reveals the Evolutionary History of the Nuclear Pore Complex. PLoS Biol 2016; 14:e1002365; PMID:26891179; https://doi.org/10.1371/journal.pbio.1002365 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
