The distribution of runs of homozygosity and selection signatures in six commercial meat sheep breeds
- PMID: 28463982
- PMCID: PMC5413029
- DOI: 10.1371/journal.pone.0176780
The distribution of runs of homozygosity and selection signatures in six commercial meat sheep breeds
Abstract
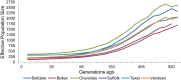
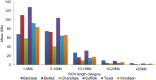
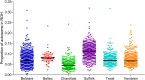
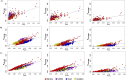
Domestication and the subsequent selection of animals for either economic or morphological features can leave a variety of imprints on the genome of a population. Genomic regions subjected to high selective pressures often show reduced genetic diversity and frequent runs of homozygosity (ROH). Therefore, the objective of the present study was to use 42,182 autosomal SNPs to identify genomic regions in 3,191 sheep from six commercial breeds subjected to selection pressure and to quantify the genetic diversity within each breed using ROH. In addition, the historical effective population size of each breed was also estimated and, in conjunction with ROH, was used to elucidate the demographic history of the six breeds. ROH were common in the autosomes of animals in the present study, but the observed breed differences in patterns of ROH length and burden suggested differences in breed effective population size and recent management. ROH provided a sufficient predictor of the pedigree inbreeding coefficient, with an estimated correlation between both measures of 0.62. Genomic regions under putative selection were identified using two complementary algorithms; the fixation index and hapFLK. The identified regions under putative selection included candidate genes associated with skin pigmentation, body size and muscle formation; such characteristics are often sought after in modern-day breeding programs. These regions of selection frequently overlapped with high ROH regions both within and across breeds. Multiple yet uncharacterised genes also resided within putative regions of selection. This further substantiates the need for a more comprehensive annotation of the sheep genome as these uncharacterised genes may contribute to traits of interest in the animal sciences. Despite this, the regions identified as under putative selection in the current study provide an insight into the mechanisms leading to breed differentiation and genetic variation in meat production.
Conflict of interest statement
Figures


Similar articles
-
Genomic insights into runs of homozygosity, effective population size and selection signatures in Iranian meat and dairy sheep breeds.PLoS One. 2025 Jun 11;20(6):e0323328. doi: 10.1371/journal.pone.0323328. eCollection 2025. PLoS One. 2025. PMID: 40498690 Free PMC article.
-
Genome-Wide Runs of Homozygosity, Effective Population Size, and Detection of Positive Selection Signatures in Six Chinese Goat Breeds.Genes (Basel). 2019 Nov 17;10(11):938. doi: 10.3390/genes10110938. Genes (Basel). 2019. PMID: 31744198 Free PMC article.
-
Runs of homozygosity analysis of South African sheep breeds from various production systems investigated using OvineSNP50k data.BMC Genomics. 2021 Jan 6;22(1):7. doi: 10.1186/s12864-020-07314-2. BMC Genomics. 2021. PMID: 33407115 Free PMC article.
-
Runs of homozygosity: current knowledge and applications in livestock.Anim Genet. 2017 Jun;48(3):255-271. doi: 10.1111/age.12526. Epub 2016 Dec 1. Anim Genet. 2017. PMID: 27910110 Review.
-
Genome-wide selection signatures detection in Shanghai Holstein cattle population identified genes related to adaption, health and reproduction traits.BMC Genomics. 2021 Oct 15;22(1):747. doi: 10.1186/s12864-021-08042-x. BMC Genomics. 2021. PMID: 34654366 Free PMC article. Review.
Cited by
-
The distribution of runs of homozygosity in the genome of river and swamp buffaloes reveals a history of adaptation, migration and crossbred events.Genet Sel Evol. 2021 Feb 27;53(1):20. doi: 10.1186/s12711-021-00616-3. Genet Sel Evol. 2021. PMID: 33639853 Free PMC article.
-
Genome-wide scans for signatures of selection in Mangalarga Marchador horses using high-throughput SNP genotyping.BMC Genomics. 2021 Oct 14;22(1):737. doi: 10.1186/s12864-021-08053-8. BMC Genomics. 2021. PMID: 34645387 Free PMC article.
-
Genomic consequences of a century of inbreeding and isolation in the Danish wild boar population.Evol Appl. 2022 May 17;15(6):954-966. doi: 10.1111/eva.13385. eCollection 2022 Jun. Evol Appl. 2022. PMID: 35782012 Free PMC article.
-
Assessing runs of Homozygosity: a comparison of SNP Array and whole genome sequence low coverage data.BMC Genomics. 2018 Jan 30;19(1):106. doi: 10.1186/s12864-018-4489-0. BMC Genomics. 2018. PMID: 29378520 Free PMC article.
-
Uncovering genomic diversity and signatures of selection in red Angus × Chinese red steppe crossbred cattle population.Sci Rep. 2025 Apr 15;15(1):12977. doi: 10.1038/s41598-025-98346-9. Sci Rep. 2025. PMID: 40234714 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

