Neuropeptide S (NPS) variants modify the signaling and risk effects of NPS Receptor 1 (NPSR1) variants in asthma
- PMID: 28463995
- PMCID: PMC5413018
- DOI: 10.1371/journal.pone.0176568
Neuropeptide S (NPS) variants modify the signaling and risk effects of NPS Receptor 1 (NPSR1) variants in asthma
Abstract
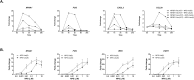
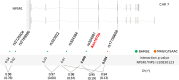
Single nucleotide polymorphisms (SNPs) close to the gain-of-function substitution, Asn(107)Ile (rs324981, A>T), in Neuropeptide S Receptor 1 (NPSR1) have been associated with asthma. Furthermore, a functional SNP (rs4751440, G>C) in Neuropeptide S (NPS) encodes a Val(6)Leu substitution on the mature peptide that results in reduced bioactivity. We sought to examine the effects of different combinations of these NPS and NPSR1 variants on downstream signaling and genetic risk of asthma. In transfected cells, the magnitude of NPSR1-induced activation of cAMP/PKA signal transduction pathways and downstream gene expression was dependent on the combination of the NPS and NPSR1 variants with NPS-Val(6)/NPSR1-Ile(107) resulting in strongest and NPS-Leu(6)/NPSR1-Asn(107) in weakest effects, respectively. One or two copies of the NPS-Leu(6) (rs4751440) were associated with physician-diagnosed childhood asthma (OR: 0.67, 95%CI 0.49-0.92, p = 0.01) and together with two other linked NPS variants (rs1931704 and rs10830123) formed a protective haplotype (p = 0.008) in the Swedish birth cohort BAMSE (2033 children). NPS rs10830123 showed epistasis with NPSR1 rs324981 encoding Asn(107)Ile (p = 0.009) in BAMSE and with the linked NPSR1 rs17199659 (p = 0.005) in the German MAGIC/ISAAC II cohort (1454 children). In conclusion, NPS variants modify asthma risk and should be considered in genetic association studies of NPSR1 with asthma and other complex diseases.
Conflict of interest statement
Figures



References
-
- Xu YL, Reinscheid RK, Huitron-Resendiz S, Clark SD, Wang Z, Lin SH, et al. Neuropeptide S: a neuropeptide promoting arousal and anxiolytic-like effects. Neuron. 2004. August 19;43(4):487–97. doi: 10.1016/j.neuron.2004.08.005 - DOI - PubMed
-
- Okamura N, Reinscheid RK. Neuropeptide S: a novel modulator of stress and arousal. Stress. 2007. August;10(3):221–6. doi: 10.1080/10253890701248673 - DOI - PubMed
-
- Guerrini R, Salvadori S, Rizzi A, Regoli D, Calo G. Neurobiology, pharmacology, and medicinal chemistry of neuropeptide S and its receptor. Medicinal research reviews. 2010. September;30(5):751–77. doi: 10.1002/med.20180 - DOI - PubMed
-
- Pape HC, Jungling K, Seidenbecher T, Lesting J, Reinscheid RK. Neuropeptide S: a transmitter system in the brain regulating fear and anxiety. Neuropharmacology. 2010. January;58(1):29–34. doi: 10.1016/j.neuropharm.2009.06.001 - DOI - PMC - PubMed
-
- Laitinen T, Polvi A, Rydman P, Vendelin J, Pulkkinen V, Salmikangas P, et al. Characterization of a common susceptibility locus for asthma-related traits. Science. 2004. April 9;304(5668):300–4. doi: 10.1126/science.1090010 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

