A high quality assembly of the Nile Tilapia (Oreochromis niloticus) genome reveals the structure of two sex determination regions
- PMID: 28464822
- PMCID: PMC5414186
- DOI: 10.1186/s12864-017-3723-5
A high quality assembly of the Nile Tilapia (Oreochromis niloticus) genome reveals the structure of two sex determination regions
Abstract
Background: Tilapias are the second most farmed fishes in the world and a sustainable source of food. Like many other fish, tilapias are sexually dimorphic and sex is a commercially important trait in these fish. In this study, we developed a significantly improved assembly of the tilapia genome using the latest genome sequencing methods and show how it improves the characterization of two sex determination regions in two tilapia species.
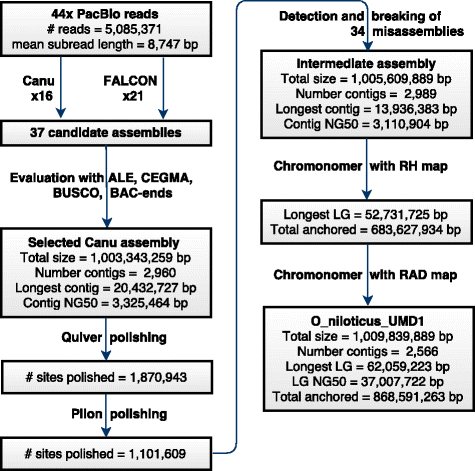
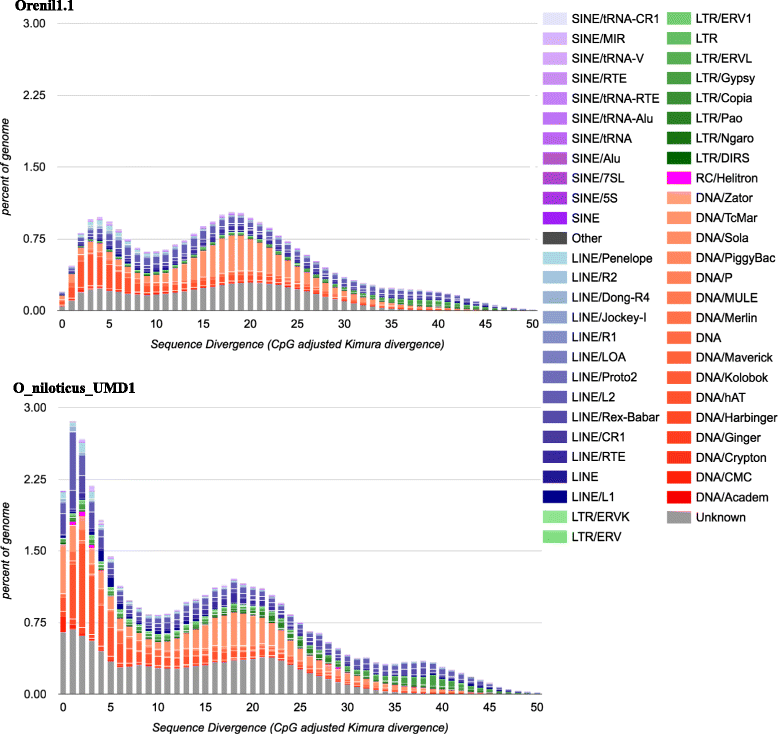
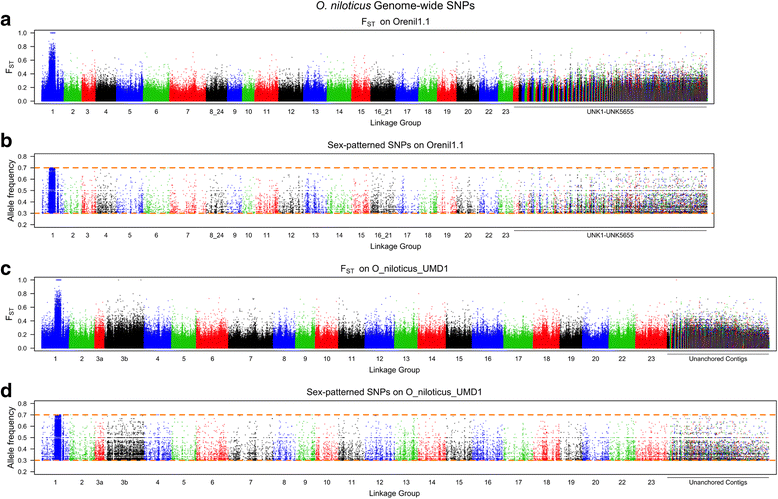
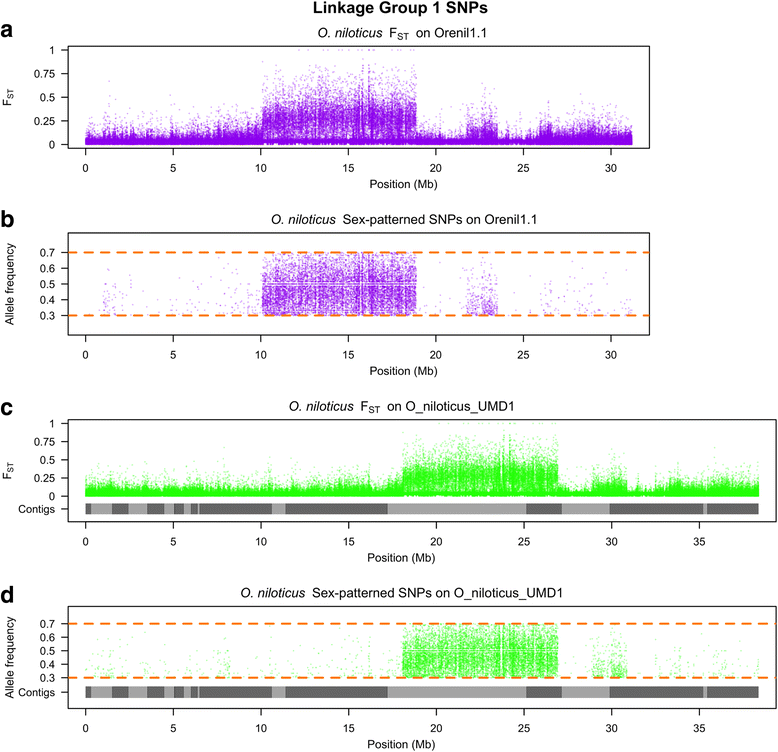
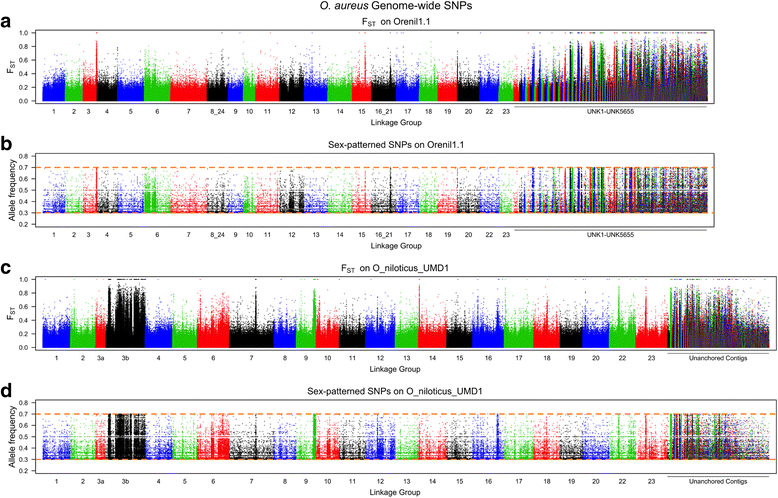
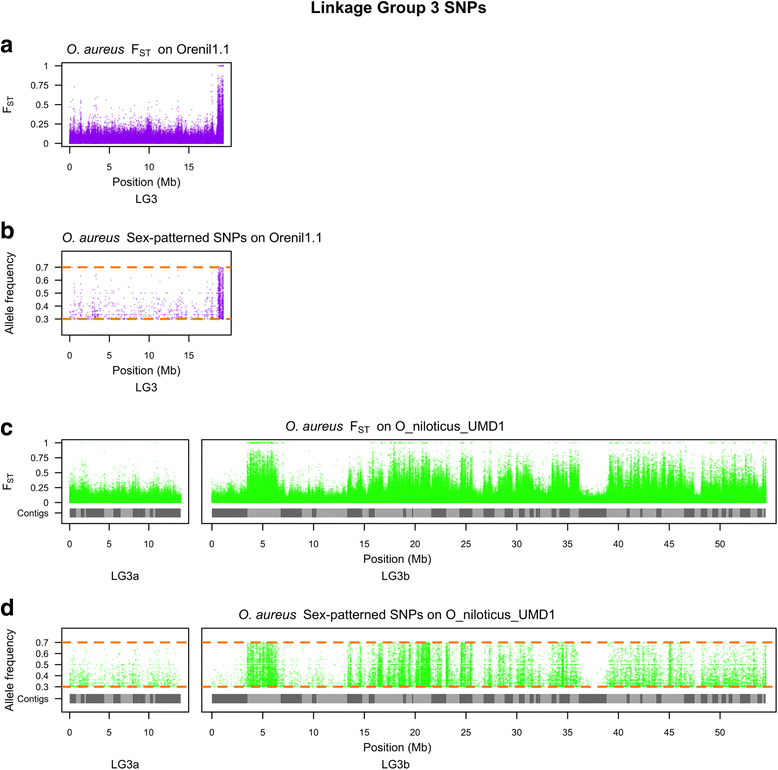
Results: A homozygous clonal XX female Nile tilapia (Oreochromis niloticus) was sequenced to 44X coverage using Pacific Biosciences (PacBio) SMRT sequencing. Dozens of candidate de novo assemblies were generated and an optimal assembly (contig NG50 of 3.3Mbp) was selected using principal component analysis of likelihood scores calculated from several paired-end sequencing libraries. Comparison of the new assembly to the previous O. niloticus genome assembly reveals that recently duplicated portions of the genome are now well represented. The overall number of genes in the new assembly increased by 27.3%, including a 67% increase in pseudogenes. The new tilapia genome assembly correctly represents two recent vasa gene duplication events that have been verified with BAC sequencing. At total of 146Mbp of additional transposable element sequence are now assembled, a large proportion of which are recent insertions. Large centromeric satellite repeats are assembled and annotated in cichlid fish for the first time. Finally, the new assembly identifies the long-range structure of both a ~9Mbp XY sex determination region on LG1 in O. niloticus, and a ~50Mbp WZ sex determination region on LG3 in the related species O. aureus.
Conclusions: This study highlights the use of long read sequencing to correctly assemble recent duplications and to characterize repeat-filled regions of the genome. The study serves as an example of the need for high quality genome assemblies and provides a framework for identifying sex determining genes in tilapia and related fish species.
Keywords: Aquaculture; Gene duplication; Genome assembly; Pacific Biosciences SMRT sequencing; Sex chromosome; Sex determination; Tilapia; Transposable elements.
Figures

Similar articles
-
Chromosome-scale assemblies reveal the structural evolution of African cichlid genomes.Gigascience. 2019 Apr 1;8(4):giz030. doi: 10.1093/gigascience/giz030. Gigascience. 2019. PMID: 30942871 Free PMC article.
-
Fine Mapping Using Whole-Genome Sequencing Confirms Anti-Müllerian Hormone as a Major Gene for Sex Determination in Farmed Nile Tilapia (Oreochromis niloticus L.).G3 (Bethesda). 2019 Oct 7;9(10):3213-3223. doi: 10.1534/g3.119.400297. G3 (Bethesda). 2019. PMID: 31416805 Free PMC article.
-
Sex determination in the GIFT strain of tilapia is controlled by a locus in linkage group 23.BMC Genet. 2020 Apr 29;21(1):49. doi: 10.1186/s12863-020-00853-3. BMC Genet. 2020. PMID: 32349678 Free PMC article.
-
A microsatellite-based linkage map of salt tolerant tilapia (Oreochromis mossambicus x Oreochromis spp.) and mapping of sex-determining loci.BMC Genomics. 2013 Jan 28;14:58. doi: 10.1186/1471-2164-14-58. BMC Genomics. 2013. PMID: 23356773 Free PMC article.
-
Integrative analysis of miRNA-mRNA expression in the brain during high temperature-induced masculinization of female Nile tilapia (Oreochromis niloticus).Genomics. 2024 May;116(3):110856. doi: 10.1016/j.ygeno.2024.110856. Epub 2024 May 10. Genomics. 2024. PMID: 38734154 Review.
Cited by
-
Disagreement in FST estimators: A case study from sex chromosomes.Mol Ecol Resour. 2020 Nov;20(6):1517-1525. doi: 10.1111/1755-0998.13210. Epub 2020 Jul 6. Mol Ecol Resour. 2020. PMID: 32543001 Free PMC article.
-
Genome assemblies for Chromidotilapia guntheri (Teleostei: Cichlidae) identify a novel candidate gene for vertebrate sex determination, RIN3.Front Genet. 2024 Aug 16;15:1447628. doi: 10.3389/fgene.2024.1447628. eCollection 2024. Front Genet. 2024. PMID: 39221227 Free PMC article.
-
The coincidence of ecological opportunity with hybridization explains rapid adaptive radiation in Lake Mweru cichlid fishes.Nat Commun. 2019 Dec 3;10(1):5391. doi: 10.1038/s41467-019-13278-z. Nat Commun. 2019. PMID: 31796733 Free PMC article.
-
Whole-Genome Sequencing and Genome-Wide Studies of Spiny Head Croaker (Collichthys lucidus) Reveals Potential Insights for Well-Developed Otoliths in the Family Sciaenidae.Front Genet. 2021 Sep 30;12:730255. doi: 10.3389/fgene.2021.730255. eCollection 2021. Front Genet. 2021. PMID: 34659355 Free PMC article.
-
Comprehensive evaluation of non-hybrid genome assembly tools for third-generation PacBio long-read sequence data.Brief Bioinform. 2019 May 21;20(3):866-876. doi: 10.1093/bib/bbx147. Brief Bioinform. 2019. PMID: 29112696 Free PMC article.
References
-
- FAO. World Review of Fisheries and Aquaculture. 2012. Available from: http://www.fao.org/docrep/016/i2727e/i2727e01.pdf.
-
- FAO. Cultured Aquatic Species Information Programme. In: FAO Fisheries and Aquaculture Department. Available from: http://www.fao.org/fishery/culturedspecies/Oreochromis_niloticus/en.
-
- Mair GC, Abucay JS, Skibinski DOF, Abella TA, Beardmore JA. Genetic manipulation of sex ratio for the large-scale production of all-male tilapia Oreochromis niloticus. Can J Fish Aquat Sci. 1997;54(2):396–404. doi: 10.1139/f96-282. - DOI
-
- Hickling C. The Malacca tilapia hybrids. J Genet. 1960;57:1–10. doi: 10.1007/BF02985334. - DOI
-
- Little D, Hulata G. Strategies for tilapia seed production. In: Beveridge M, McAndrew B, editors. Tilapias: biology and exploitation. 2000. pp. 267–326.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

