Mutant IDH1 regulates the tumor-associated immune system in gliomas
- PMID: 28465358
- PMCID: PMC5435890
- DOI: 10.1101/gad.294991.116
Mutant IDH1 regulates the tumor-associated immune system in gliomas
Abstract
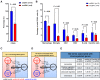
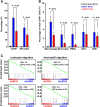
Gliomas harboring mutations in isocitrate dehydrogenase 1/2 (IDH1/2) have the CpG island methylator phenotype (CIMP) and significantly longer patient survival time than wild-type IDH1/2 (wtIDH1/2) tumors. Although there are many factors underlying the differences in survival between these two tumor types, immune-related differences in cell content are potentially important contributors. In order to investigate the role of IDH mutations in immune response, we created a syngeneic pair mouse model for mutant IDH1 (muIDH1) and wtIDH1 gliomas and demonstrated that muIDH1 mice showed many molecular and clinical similarities to muIDH1 human gliomas, including a 100-fold higher concentration of 2-hydroxygluratate (2-HG), longer survival time, and higher CpG methylation compared with wtIDH1. Also, we showed that IDH1 mutations caused down-regulation of leukocyte chemotaxis, resulting in repression of the tumor-associated immune system. Given that significant infiltration of immune cells such as macrophages, microglia, monocytes, and neutrophils is linked to poor prognosis in many cancer types, these reduced immune infiltrates in muIDH1 glioma tumors may contribute in part to the differences in aggressiveness of the two glioma types.
Keywords: IDH mutation; glioma; immuno-oncology.
© 2017 Amankulor et al.; Published by Cold Spring Harbor Laboratory Press.
Figures




References
-
- Badie B, Schartner JM. 2000. Flow cytometric characterization of tumor-associated macrophages in experimental gliomas. Neurosurgery 46: 957–961. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
