Ethylene-Inhibited Jasmonic Acid Biosynthesis Promotes Mesocotyl/Coleoptile Elongation of Etiolated Rice Seedlings
- PMID: 28465411
- PMCID: PMC5466032
- DOI: 10.1105/tpc.16.00981
Ethylene-Inhibited Jasmonic Acid Biosynthesis Promotes Mesocotyl/Coleoptile Elongation of Etiolated Rice Seedlings
Abstract
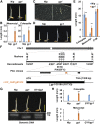
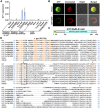
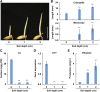
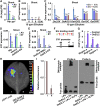
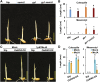
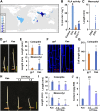
Elongation of the mesocotyl and coleoptile facilitates the emergence of rice (Oryza sativa) seedlings from soil and is affected by various genetic and environment factors. The regulatory mechanism underlying this process remains largely unclear. Here, we examined the regulation of mesocotyl and coleoptile growth by characterizing a gaoyao1 (gy1) mutant that exhibits a longer mesocotyl and longer coleoptile than its original variety of rice. GY1 was identified through map-based cloning and encodes a PLA1-type phospholipase that localizes in chloroplasts. GY1 functions at the initial step of jasmonic acid (JA) biosynthesis to repress mesocotyl and coleoptile elongation in etiolated rice seedlings. Ethylene inhibits the expression of GY1 and other genes in the JA biosynthesis pathway to reduce JA levels and enhance mesocotyl and coleoptile growth by promoting cell elongation. Genetically, GY1 acts downstream of the OsEIN2-mediated ethylene signaling pathway to regulate mesocotyl/coleoptile growth. Through analysis of the resequencing data from 3000 rice accessions, we identified a single natural variation of the GY1 gene, GY1376T , which contributes to mesocotyl elongation in rice varieties. Our study reveals novel insights into the regulatory mechanism of mesocotyl/coleoptile elongation and should have practical applications in rice breeding programs.
© 2017 American Society of Plant Biologists. All rights reserved.
Figures


Similar articles
-
Ethylene-induced inhibition of root growth requires abscisic acid function in rice (Oryza sativa L.) seedlings.PLoS Genet. 2014 Oct 16;10(10):e1004701. doi: 10.1371/journal.pgen.1004701. eCollection 2014 Oct. PLoS Genet. 2014. PMID: 25330236 Free PMC article.
-
An orchestrated ethylene-gibberellin signaling cascade contributes to mesocotyl elongation and emergence of rice direct seeding.J Integr Plant Biol. 2024 Jul;66(7):1427-1439. doi: 10.1111/jipb.13671. Epub 2024 May 15. J Integr Plant Biol. 2024. PMID: 38751025
-
OsEIL1 and OsEIL2, two master regulators of rice ethylene signaling, promote the expression of ROS scavenging genes to facilitate coleoptile elongation and seedling emergence from soil.Plant Commun. 2024 Mar 11;5(3):100771. doi: 10.1016/j.xplc.2023.100771. Epub 2023 Nov 22. Plant Commun. 2024. PMID: 37994014 Free PMC article.
-
Ethylene-mediated regulation of coleoptile elongation in rice seedlings.Plant Cell Environ. 2023 Apr;46(4):1060-1074. doi: 10.1111/pce.14492. Epub 2022 Dec 2. Plant Cell Environ. 2023. PMID: 36397123 Review.
-
Mesocotyl elongation, an essential trait for dry-seeded rice (Oryza sativa L.): a review of physiological and genetic basis.Planta. 2019 Dec 4;251(1):27. doi: 10.1007/s00425-019-03322-z. Planta. 2019. PMID: 31802259 Review.
Cited by
-
Transcriptome Analysis Revealed the Key Genes and Pathways Involved in Seed Germination of Maize Tolerant to Deep-Sowing.Plants (Basel). 2022 Jan 28;11(3):359. doi: 10.3390/plants11030359. Plants (Basel). 2022. PMID: 35161340 Free PMC article.
-
Rice EIL1 interacts with OsIAAs to regulate auxin biosynthesis mediated by the tryptophan aminotransferase MHZ10/OsTAR2 during root ethylene responses.Plant Cell. 2022 Oct 27;34(11):4366-4387. doi: 10.1093/plcell/koac250. Plant Cell. 2022. PMID: 35972379 Free PMC article.
-
Integrating Association Mapping, Linkage Mapping, Fine Mapping with RNA Seq Conferring Seedling Vigor Improvement for Successful Crop Establishment in Deep Sown Direct-Seeded Rice.Rice (N Y). 2023 Oct 17;16(1):46. doi: 10.1186/s12284-023-00665-w. Rice (N Y). 2023. PMID: 37848638 Free PMC article.
-
Ethylene Signaling Facilitates Plant Adaption to Physical Barriers.Front Plant Sci. 2021 Jul 29;12:697988. doi: 10.3389/fpls.2021.697988. eCollection 2021. Front Plant Sci. 2021. PMID: 34394151 Free PMC article. Review.
-
Enhancing sweet sorghum emergence and stress resilience in saline-alkaline soils through ABA seed priming: insights into hormonal and metabolic reprogramming.BMC Genomics. 2025 Mar 12;26(1):241. doi: 10.1186/s12864-025-11420-4. BMC Genomics. 2025. PMID: 40075293 Free PMC article.
References
-
- Alonso J.M., Hirayama T., Roman G., Nourizadeh S., Ecker J.R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152. - PubMed
-
- Bannenberg G., Martínez M., Hamberg M., Castresana C. (2009). Diversity of the enzymatic activity in the lipoxygenase gene family of Arabidopsis thaliana. Lipids 44: 85–95. - PubMed
-
- Biswas K.K., Neumann R., Haga K., Yatoh O., Iino M. (2003). Photomorphogenesis of rice seedlings: a mutant impaired in phytochrome-mediated inhibition of coleoptile growth. Plant Cell Physiol. 44: 242–254. - PubMed
-
- Bleecker A.B., Kende H. (2000). Ethylene: a gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16: 1–18. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

