Gene expression profiling describes the genetic regulation of Meloidogyne arenaria resistance in Arachis hypogaea and reveals a candidate gene for resistance
- PMID: 28465503
- PMCID: PMC5430994
- DOI: 10.1038/s41598-017-00971-6
Gene expression profiling describes the genetic regulation of Meloidogyne arenaria resistance in Arachis hypogaea and reveals a candidate gene for resistance
Abstract
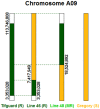
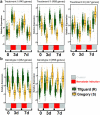
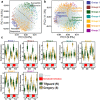
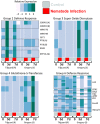
Resistance to root-knot nematode was introgressed into cultivated peanut Arachis hypogaea from a wild peanut relative, A. cardenasii and previously mapped to chromosome A09. The highly resistant recombinant inbred RIL 46 and moderately resistant RIL 48 were selected from a population with cv. Gregory (susceptible) and Tifguard (resistant) as female and male parents, respectively. RNA-seq analysis was performed on these four genotypes using root tissue harvested from root-knot nematode infected plants at 0, 3, 7 days after inoculation. Differential gene expression analysis provides evidence that root-knot nematodes modulate biological pathways involved in plant hormone, defense, cell signaling, cytoskeleton and cell wall metabolism in a susceptible reaction. Corresponding to resistance reaction, an effector-induced-immune response mediated by an R-gene was identified in Tifguard. Mapping of the introgressed region indicated that 92% of linkage group A09 was of A. cardenasii origin in Tifguard. RIL46 and RIL 48 possessed 3.6% and 83.5% of the introgression on A09, respectively. Within the small introgressed region carried by RIL 46, a constitutively expressed TIR-NBS-LRR gene was identified as the candidate for nematode resistance. Potential defense responsive pathways include effector endocytosis through clathrin-coated vesicle trafficking, defense signaling through membrane lipid metabolism and mucilage production.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Minton, N. A. & Baujard, P. Nematode parasites of peanut. Plant parasitic nematodes in subtropical and tropical agriculture. M. Luc, R. A. Sikora and J. Bridge. Wallingford, UK, CABl, 285–320 (1990).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

