Strategies in 'snake venomics' aiming at an integrative view of compositional, functional, and immunological characteristics of venoms
- PMID: 28465677
- PMCID: PMC5408369
- DOI: 10.1186/s40409-017-0117-8
Strategies in 'snake venomics' aiming at an integrative view of compositional, functional, and immunological characteristics of venoms
Abstract
This work offers a general overview on the evolving strategies for the proteomic analysis of snake venoms, and discusses how these may be combined through diverse experimental approaches with the goal of achieving a more comprehensive knowledge on the compositional, toxic, and immunological characteristics of venoms. Some recent developments in this field are summarized, highlighting how strategies have evolved from the mere cataloguing of venom components (proteomics/venomics), to a broader exploration of their immunological (antivenomics) and functional (toxicovenomics) characteristics. Altogether, the combination of these complementary strategies is helping to build a wider, more integrative view of the life-threatening protein cocktails produced by venomous snakes, responsible for thousands of deaths every year.
Keywords: Antivenomics; Proteomics; Snake venoms; Toxicovenomics; Venomics.
Figures
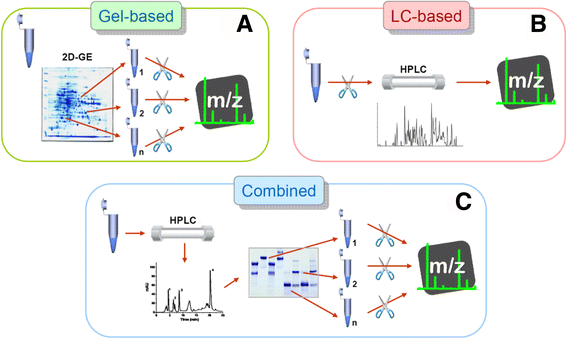
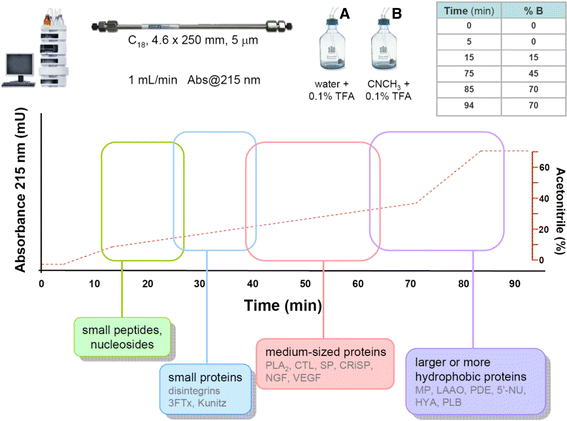
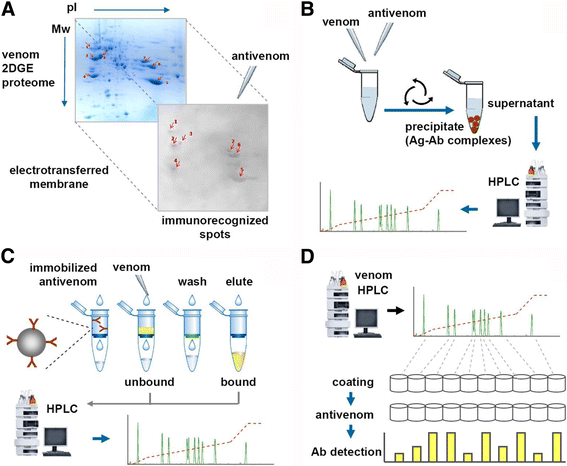
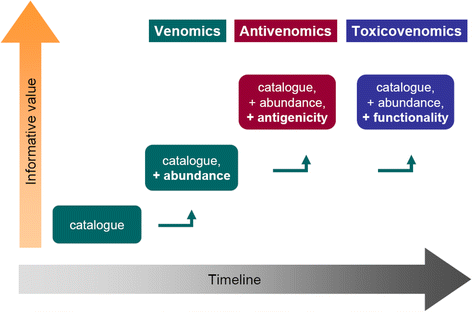
References
-
- Garfield E. Research and immunotherapy are taking the bite out of venom. Curr Contents. 1988;5(11):29–38.
-
- Mackessy SP, (editor). Handbook of Venoms and Toxins of Reptiles. CRC Press, Taylor & Francis Group, Boca Raton, FL, USA; 2009. ISBN 978-0849391651.
-
- Fry B, editor. Venomous Reptiles and Their Toxins: Evolution, Pathophysiology and Biodiscovery. Oxford: Oxford University Press; 2015.
-
- Fry BG, Sunagar K, Casewell NR, Kochva E, Roelants K, Jackson TNW, et al. The origin and evolution of the Toxicofera reptile venom system. In: Brian F, editor. Venomous Reptiles and their Toxins. Oxford: Oxford University Press; 2015. p. 1-31.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

