Evolutionary history of LTR-retrotransposons among 20 Drosophila species
- PMID: 28465726
- PMCID: PMC5408442
- DOI: 10.1186/s13100-017-0090-3
Evolutionary history of LTR-retrotransposons among 20 Drosophila species
Abstract
Background: The presence of transposable elements (TEs) in genomes is known to explain in part the variations of genome sizes among eukaryotes. Even among closely related species, the variation of TE amount may be striking, as for example between the two sibling species, Drosophila melanogaster and D. simulans. However, not much is known concerning the TE content and dynamics among other Drosophila species. The sequencing of several Drosophila genomes, covering the two subgenus Sophophora and Drosophila, revealed a large variation of the repeat content among these species but no much information is known concerning their precise TE content. The identification of some consensus sequences of TEs from the various sequenced Drosophila species allowed to get an idea concerning their variety in term of diversity of superfamilies but the used classification remains very elusive and ambiguous.
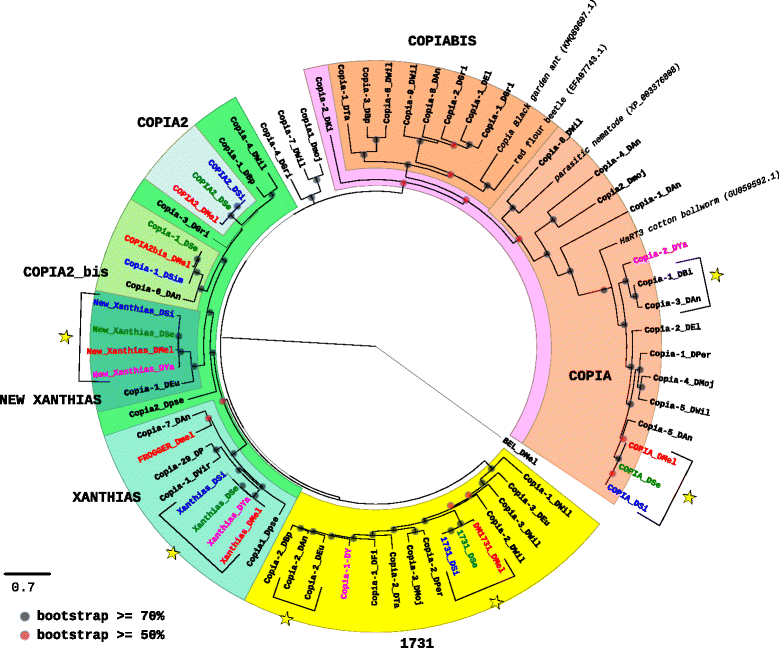
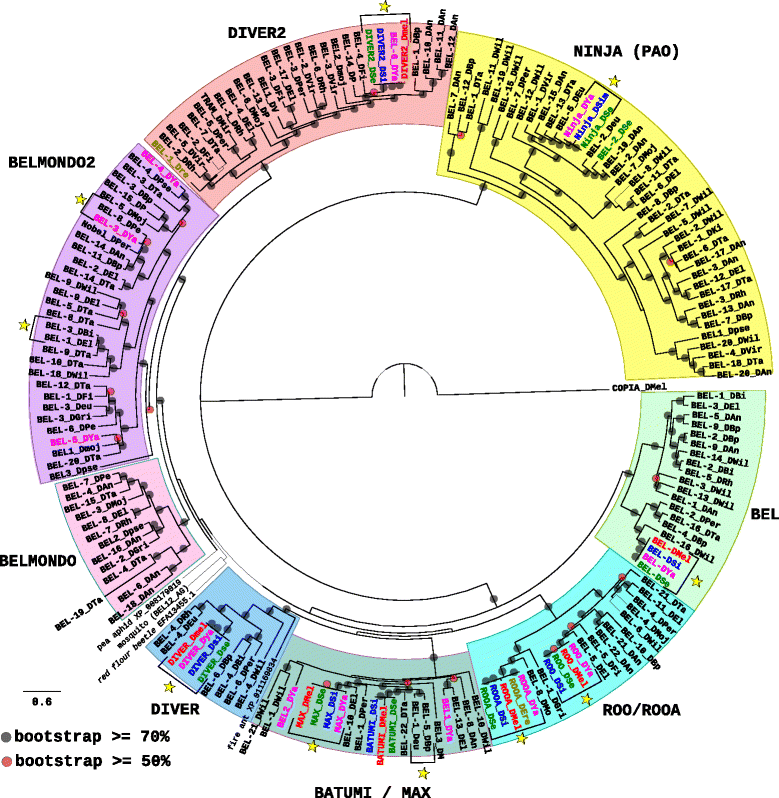
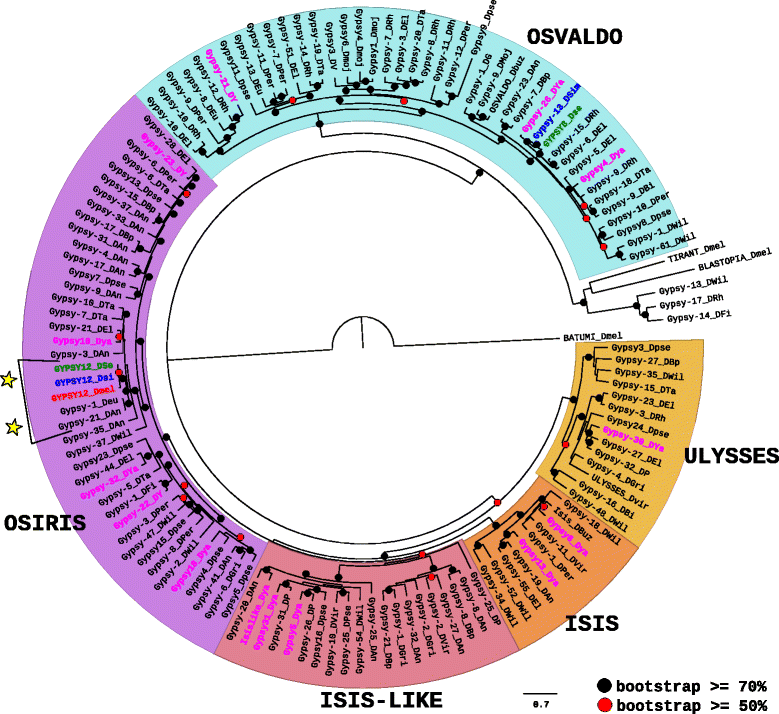
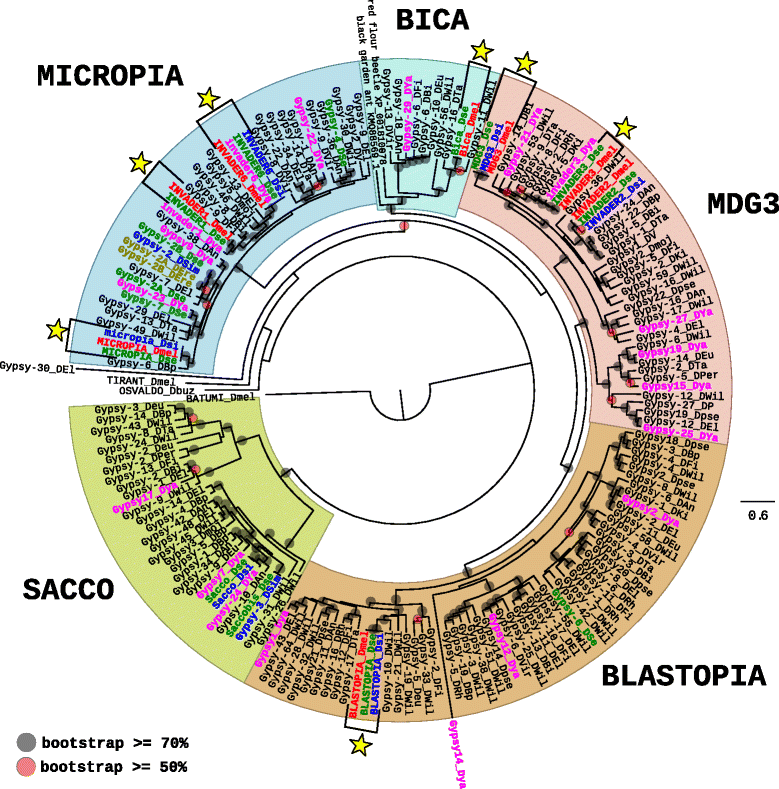
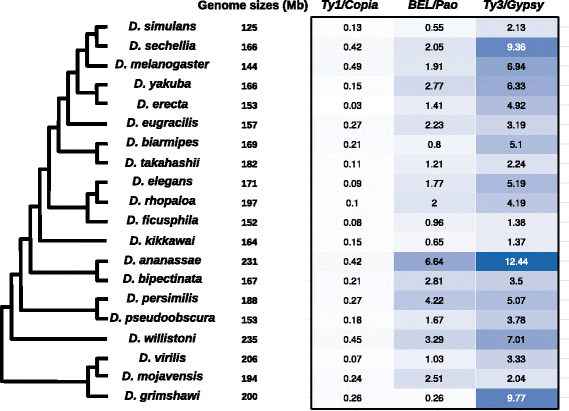
Results: We choose to focus on LTR-retrotransposons because they represent the most widely represented class of TEs in the Drosophila genomes. In this work, we describe for the first time the phylogenetic relationship of each LTR-retrotransposon family described in 20 Drosophila species, compute their proportion in their respective genomes and identify several new cases of horizontal transfers.
Conclusion: All these results allow us to have a clearer view on the evolutionary history of LTR retrotransposons among Drosophila that seems to be mainly driven by vertical transmissions although the implications of horizontal transfers, losses and intra-specific diversification are clearly also at play.
Keywords: Drosophila; Horizontal transfer; LTR-retrotransposons; Transposable element dynamics.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

