Enzyme discovery beyond homology: a unique hydroxynitrile lyase in the Bet v1 superfamily
- PMID: 28466867
- PMCID: PMC5413884
- DOI: 10.1038/srep46738
Enzyme discovery beyond homology: a unique hydroxynitrile lyase in the Bet v1 superfamily
Erratum in
-
Erratum: Enzyme discovery beyond homology: a unique hydroxynitrile lyase in the Bet v1 superfamily.Sci Rep. 2017 May 26;7:46834. doi: 10.1038/srep46834. Sci Rep. 2017. PMID: 28548662 Free PMC article.
Abstract
Homology and similarity based approaches are most widely used for the identification of new enzymes for biocatalysis. However, they are not suitable to find truly novel scaffolds with a desired function and this averts options and diversity. Hydroxynitrile lyases (HNLs) are an example of non-homologous isofunctional enzymes for the synthesis of chiral cyanohydrins. Due to their convergent evolution, finding new representatives is challenging. Here we show the discovery of unique HNL enzymes from the fern Davallia tyermannii by coalescence of transcriptomics, proteomics and enzymatic screening. It is the first protein with a Bet v1-like protein fold exhibiting HNL activity, and has a new catalytic center, as shown by protein crystallography. Biochemical properties of D. tyermannii HNLs open perspectives for the development of a complementary class of biocatalysts for the stereoselective synthesis of cyanohydrins. This work shows that systematic integration of -omics data facilitates discovery of enzymes with unpredictable sequences and helps to extend our knowledge about enzyme diversity.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
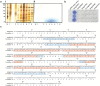

 ; DtHNL2
; DtHNL2  ; DtHNL3
; DtHNL3  ; DtHNL4
; DtHNL4  . Grey dashed lines indicate the spontaneous degradation of racemic mandelonitrile in a negative control reaction without enzyme addition (background reaction). Standard enzymatic assay was performed by monitoring benzaldehyde formation at 280 nm. Values were obtained from the average of a minimal of two and a maximum of three independent samples, each of which is the average of two or three technical replicates. Standard deviations are within the 20% threshold (or 25% for temperature profile). For clarity, error bars have been omitted. (a) pH profile. Relative activity of DtHNL isoenzymes at different pH values from 2.0 to 7.0. The assay was performed in HCl-potassium chloride buffer (filled symbols), or sodium citrate-phosphate buffer (empty symbols). Activity of DtHNL1 and 2 at pH 7.0 is not depicted due to high standard deviations. (b) Temperature profile. Relative activity of DtHNL isoenzymes at different temperatures from 10 to 50 °C. The assay was performed at pH 5.0. Omitted points are due to high standard deviations. Enzyme stability at pH 2.5 (c) and at pH 4.0 (d). Activity after incubation of DtHNL isoenzymes at pH 2.5 or 4.0, respectively, and 8 °C. Relative activity is based on the activity before incubation.
. Grey dashed lines indicate the spontaneous degradation of racemic mandelonitrile in a negative control reaction without enzyme addition (background reaction). Standard enzymatic assay was performed by monitoring benzaldehyde formation at 280 nm. Values were obtained from the average of a minimal of two and a maximum of three independent samples, each of which is the average of two or three technical replicates. Standard deviations are within the 20% threshold (or 25% for temperature profile). For clarity, error bars have been omitted. (a) pH profile. Relative activity of DtHNL isoenzymes at different pH values from 2.0 to 7.0. The assay was performed in HCl-potassium chloride buffer (filled symbols), or sodium citrate-phosphate buffer (empty symbols). Activity of DtHNL1 and 2 at pH 7.0 is not depicted due to high standard deviations. (b) Temperature profile. Relative activity of DtHNL isoenzymes at different temperatures from 10 to 50 °C. The assay was performed at pH 5.0. Omitted points are due to high standard deviations. Enzyme stability at pH 2.5 (c) and at pH 4.0 (d). Activity after incubation of DtHNL isoenzymes at pH 2.5 or 4.0, respectively, and 8 °C. Relative activity is based on the activity before incubation.

Similar articles
-
Production of Hydroxynitrile Lyase from Davallia tyermannii (DtHNL) in Komagataella phaffii and Its Immobilization as a CLEA to Generate a Robust Biocatalyst.Chembiochem. 2018 Feb 16;19(4):312-316. doi: 10.1002/cbic.201700419. Epub 2017 Dec 11. Chembiochem. 2018. PMID: 29131473
-
Modern Approaches to Discovering New Hydroxynitrile Lyases for Biocatalysis.Chembiochem. 2017 Jan 17;18(2):152-160. doi: 10.1002/cbic.201600495. Epub 2016 Nov 29. Chembiochem. 2017. PMID: 27898188 Review.
-
Discovery and molecular and biocatalytic properties of hydroxynitrile lyase from an invasive millipede, Chamberlinius hualienensis.Proc Natl Acad Sci U S A. 2015 Aug 25;112(34):10605-10. doi: 10.1073/pnas.1508311112. Epub 2015 Aug 10. Proc Natl Acad Sci U S A. 2015. PMID: 26261304 Free PMC article.
-
The crystal structure and catalytic mechanism of hydroxynitrile lyase from passion fruit, Passiflora edulis.FEBS J. 2018 Jan;285(2):313-324. doi: 10.1111/febs.14339. Epub 2017 Dec 7. FEBS J. 2018. PMID: 29155493
-
Potential and capabilities of hydroxynitrile lyases as biocatalysts in the chemical industry.Appl Microbiol Biotechnol. 2007 Aug;76(2):309-20. doi: 10.1007/s00253-007-1025-6. Epub 2007 Jul 3. Appl Microbiol Biotechnol. 2007. PMID: 17607575 Review.
Cited by
-
Erratum: Enzyme discovery beyond homology: a unique hydroxynitrile lyase in the Bet v1 superfamily.Sci Rep. 2017 May 26;7:46834. doi: 10.1038/srep46834. Sci Rep. 2017. PMID: 28548662 Free PMC article.
-
Synthesis of (R)-mandelic acid and (R)-mandelic acid amide by recombinant E. coli strains expressing a (R)-specific oxynitrilase and an arylacetonitrilase.Biotechnol Lett. 2021 Jan;43(1):287-296. doi: 10.1007/s10529-020-02998-8. Epub 2020 Sep 16. Biotechnol Lett. 2021. PMID: 32936375 Free PMC article.
-
Structural analysis of a ligand-triggered intermolecular disulfide switch in a major latex protein from opium poppy.Acta Crystallogr D Struct Biol. 2024 Sep 1;80(Pt 9):675-685. doi: 10.1107/S2059798324007733. Epub 2024 Aug 29. Acta Crystallogr D Struct Biol. 2024. PMID: 39207895 Free PMC article.
-
Structural characterization of Linum usitatissimum hydroxynitrile lyase: A new cyanohydrin decomposition mechanism involving a cyano-zinc complex.J Biol Chem. 2022 Mar;298(3):101650. doi: 10.1016/j.jbc.2022.101650. Epub 2022 Jan 29. J Biol Chem. 2022. PMID: 35101448 Free PMC article.
-
Progress in Stereoselective Construction of C-C Bonds Enabled by Aldolases and Hydroxynitrile Lyases.Front Bioeng Biotechnol. 2021 Apr 21;9:653682. doi: 10.3389/fbioe.2021.653682. eCollection 2021. Front Bioeng Biotechnol. 2021. PMID: 33968915 Free PMC article. Review.
References
-
- Kassim M. A. & Rumbold K. HCN production and hydroxynitrile lyase: a natural activity in plants and a renewed biotechnological interest. Biotechnol. Lett. 36, 223–8 (2014). - PubMed
-
- Dadashipour M. & Asano Y. Hydroxynitrile lyases: insights into biochemistry, discovery, and engineering. ACS Catal. 1, 1121–1149 (2011).
-
- Sharma M., Sharma N. N. & Bhalla T. C. Hydroxynitrile lyases: At the interface of biology and chemistry. Enzyme Microb. Technol. 37, 279–294 (2005).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

