PPARδ Promotes Running Endurance by Preserving Glucose
- PMID: 28467934
- PMCID: PMC5492977
- DOI: 10.1016/j.cmet.2017.04.006
PPARδ Promotes Running Endurance by Preserving Glucose
Abstract
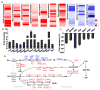
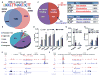
Management of energy stores is critical during endurance exercise; a shift in substrate utilization from glucose toward fat is a hallmark of trained muscle. Here we show that this key metabolic adaptation is both dependent on muscle PPARδ and stimulated by PPARδ ligand. Furthermore, we find that muscle PPARδ expression positively correlates with endurance performance in BXD mouse reference populations. In addition to stimulating fatty acid metabolism in sedentary mice, PPARδ activation potently suppresses glucose catabolism and does so without affecting either muscle fiber type or mitochondrial content. By preserving systemic glucose levels, PPARδ acts to delay the onset of hypoglycemia and extends running time by ∼100 min in treated mice. Collectively, these results identify a bifurcated PPARδ program that underlies glucose sparing and highlight the potential of PPARδ-targeted exercise mimetics in the treatment of metabolic disease, dystrophies, and, unavoidably, the enhancement of athletic performance.
Keywords: PPARδ; endurance exercise; exercise mimetics; fatty acid metabolism; glucose metabolism; muscle.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures


References
-
- Bruce CR, Hoy AJ, Turner N, Watt MJ, Allen TL, Carpenter K, Cooney GJ, Febbraio MA, Kraegen EW. Overexpression of carnitine palmitoyltransferase-1 in skeletal muscle is sufficient to enhance fatty acid oxidation and improve high-fat diet-induced insulin resistance. Diabetes. 2009;58:550–558. - PMC - PubMed
-
- Dressel U, Allen TL, Pippal JB, Rohde PR, Lau P, Muscat GE. The peroxisome proliferator-activated receptor beta/delta agonist, GW501516, regulates the expression of genes involved in lipid catabolism and energy uncoupling in skeletal muscle cells. Molecular endocrinology. 2003;17:2477–2493. - PubMed
-
- Duan SZ, Usher MG, Mortensen RM. Peroxisome proliferator-activated receptor-gamma-mediated effects in the vasculature. Circulation research. 2008;102:283–294. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

