Sampling Terrestrial Environments for Bacterial Polyketides
- PMID: 28468277
- PMCID: PMC6154731
- DOI: 10.3390/molecules22050707
Sampling Terrestrial Environments for Bacterial Polyketides
Abstract
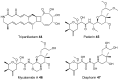
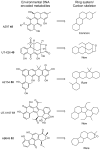
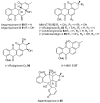
Bacterial polyketides are highly biologically active molecules that are frequently used as drugs, particularly as antibiotics and anticancer agents, thus the discovery of new polyketides is of major interest. Since the 1980s discovery of polyketides has slowed dramatically due in large part to the repeated rediscovery of known compounds. While recent scientific and technical advances have improved our ability to discover new polyketides, one key area has been under addressed, namely the distribution of polyketide-producing bacteria in the environment. Identifying environments where producing bacteria are abundant and diverse should improve our ability to discover (bioprospect) new polyketides. This review summarizes for the bioprospector the state-of-the-field in terrestrial microbial ecology. It provides insight into the scientific and technical challenges limiting the application of microbial ecology discoveries for bioprospecting and summarizes key developments in the field that will enable more effective bioprospecting. The major recent efforts by researchers to sample new environments for polyketide discovery is also reviewed and key emerging environments such as insect associated bacteria, desert soils, disease suppressive soils, and caves are highlighted. Finally strategies for taking and characterizing terrestrial samples to help maximize discovery efforts are proposed and the inclusion of non-actinomycetal bacteria in any terrestrial discovery strategy is recommended.
Keywords: bioprospecting; microbial ecology; polyketides.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

















Similar articles
-
Microbial polyketides and their roles in insect virulence: from genomics to biological functions.Nat Prod Rep. 2022 Nov 16;39(11):2008-2029. doi: 10.1039/d1np00058f. Nat Prod Rep. 2022. PMID: 35822627 Review.
-
Tafuketide, a phylogeny-guided discovery of a new polyketide from Talaromyces funiculosus Salicorn 58.Appl Microbiol Biotechnol. 2016 Jun;100(12):5323-38. doi: 10.1007/s00253-016-7311-4. Epub 2016 Jan 26. Appl Microbiol Biotechnol. 2016. PMID: 26810200
-
Antibacterial Aromatic Polyketides Incorporating the Unusual Amino Acid Enduracididine.J Nat Prod. 2019 Jan 25;82(1):35-44. doi: 10.1021/acs.jnatprod.8b00354. Epub 2019 Jan 7. J Nat Prod. 2019. PMID: 30615447
-
Engyodontochones, Antibiotic Polyketides from the Marine Fungus Engyodontium album Strain LF069.Chemistry. 2016 May 23;22(22):7452-62. doi: 10.1002/chem.201600430. Epub 2016 Apr 21. Chemistry. 2016. PMID: 27098103
-
Spirotetronate polyketides as leads in drug discovery.J Nat Prod. 2015 Mar 27;78(3):562-75. doi: 10.1021/np500757w. Epub 2014 Dec 1. J Nat Prod. 2015. PMID: 25434976 Free PMC article. Review.
Cited by
-
Diversity, Ecology, and Prevalence of Antimicrobials in Nature.Front Microbiol. 2019 Nov 14;10:2518. doi: 10.3389/fmicb.2019.02518. eCollection 2019. Front Microbiol. 2019. PMID: 31803148 Free PMC article. Review.
-
Metagenomics Unveils Posidonia oceanica "Banquettes" as a Potential Source of Novel Bioactive Compounds and Carbohydrate Active Enzymes (CAZymes).mSystems. 2021 Oct 26;6(5):e0086621. doi: 10.1128/mSystems.00866-21. Epub 2021 Sep 14. mSystems. 2021. PMID: 34519521 Free PMC article.
-
Microbiota Composition and Functional Profiling Throughout the Gastrointestinal Tract of Commercial Weaning Piglets.Microorganisms. 2019 Sep 12;7(9):343. doi: 10.3390/microorganisms7090343. Microorganisms. 2019. PMID: 31547478 Free PMC article.
-
Draft Genome Sequence of the Type Strain Streptomyces armeniacus ATCC 15676.Microbiol Resour Announc. 2018 Sep 20;7(11):e01107-18. doi: 10.1128/MRA.01107-18. eCollection 2018 Sep. Microbiol Resour Announc. 2018. PMID: 30533638 Free PMC article.
-
Isolation, Characterization, and Antibacterial Activity of Hard-to-Culture Actinobacteria from Cave Moonmilk Deposits.Antibiotics (Basel). 2018 Mar 22;7(2):28. doi: 10.3390/antibiotics7020028. Antibiotics (Basel). 2018. PMID: 29565274 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

