Regulation of Tacaribe Mammarenavirus Translation: Positive 5' and Negative 3' Elements and Role of Key Cellular Factors
- PMID: 28468879
- PMCID: PMC5487564
- DOI: 10.1128/JVI.00084-17
Regulation of Tacaribe Mammarenavirus Translation: Positive 5' and Negative 3' Elements and Role of Key Cellular Factors
Abstract
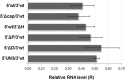
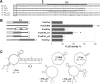
Mammarenaviruses are enveloped viruses with a bisegmented negative-stranded RNA genome that encodes the nucleocapsid protein (NP), the envelope glycoprotein precursor (GPC), the RNA polymerase (L), and a RING matrix protein (Z). Viral proteins are synthesized from subgenomic mRNAs bearing a capped 5' untranslated region (UTR) and lacking 3' poly(A) tail. We analyzed the translation strategy of Tacaribe virus (TCRV), a prototype of the New World mammarenaviruses. A virus-like transcript that carries a reporter gene in place of the NP open reading frame and transcripts bearing modified 5' and/or 3' UTR were evaluated in a cell-based translation assay. We found that the presence of the cap structure at the 5' end dramatically increases translation efficiency and that the viral 5' UTR comprises stimulatory signals while the 3' UTR,specifically the presence of a terminal C+G-rich sequence and/or a stem-loop structure, down-modulates translation. Additionally, translation was profoundly reduced in eukaryotic initiation factor (eIF) 4G-inactivated cells, whereas depletion of intracellular levels of eIF4E had less impact on virus-like mRNA translation than on a cell-like transcript. Translation efficiency was independent of NP expression or TCRV infection. Our results indicate that TCRV mRNAs are translated using a cap-dependent mechanism, whose efficiency relies on the interplay between stimulatory signals in the 5' UTR and a negative modulatory element in the 3' UTR. The low dependence on eIF4E suggests that viral mRNAs may engage yet-unknown noncanonical host factors for a cap-dependent initiation mechanism.IMPORTANCE Several members of the Arenaviridae family cause serious hemorrhagic fevers in humans. In the present report, we describe the mechanism by which Tacaribe virus, a prototypic nonpathogenic New World mammarenavirus, regulates viral mRNA translation. Our results highlight the impact of untranslated sequences and key host translation factors on this process. We propose a model that explains how viral mRNAs outcompete cellular mRNAs for the translation machinery. A better understanding of the mechanism of translation regulation of this virus can provide the bases for the rational design of new antiviral tools directed to pathogenic arenaviruses.
Keywords: arenavirus; mRNA; translation.
Copyright © 2017 American Society for Microbiology.
Figures


Similar articles
-
A four-nucleotide translation enhancer in the 3'-terminal consensus sequence of the nonpolyadenylated mRNAs of rotavirus.RNA. 2000 Jun;6(6):814-25. doi: 10.1017/s1355838200992264. RNA. 2000. PMID: 10864041 Free PMC article.
-
Control of translation by the 5'- and 3'-terminal regions of the dengue virus genome.J Virol. 2005 Jul;79(13):8303-15. doi: 10.1128/JVI.79.13.8303-8315.2005. J Virol. 2005. PMID: 15956576 Free PMC article.
-
Dengue virus utilizes a novel strategy for translation initiation when cap-dependent translation is inhibited.J Virol. 2006 Mar;80(6):2976-86. doi: 10.1128/JVI.80.6.2976-2986.2006. J Virol. 2006. PMID: 16501107 Free PMC article.
-
Translational control of mRNAs by 3'-Untranslated region binding proteins.BMB Rep. 2017 Apr;50(4):194-200. doi: 10.5483/bmbrep.2017.50.4.040. BMB Rep. 2017. PMID: 28287067 Free PMC article. Review.
-
Control of translation initiation in animals.Annu Rev Cell Dev Biol. 1998;14:399-458. doi: 10.1146/annurev.cellbio.14.1.399. Annu Rev Cell Dev Biol. 1998. PMID: 9891789 Review.
Cited by
-
Hemorrhagic Fever-Causing Arenaviruses: Lethal Pathogens and Potent Immune Suppressors.Front Immunol. 2019 Mar 13;10:372. doi: 10.3389/fimmu.2019.00372. eCollection 2019. Front Immunol. 2019. PMID: 30918506 Free PMC article. Review.
-
DDX3 suppresses type I interferons and favors viral replication during Arenavirus infection.PLoS Pathog. 2018 Jul 12;14(7):e1007125. doi: 10.1371/journal.ppat.1007125. eCollection 2018 Jul. PLoS Pathog. 2018. PMID: 30001425 Free PMC article.
-
Nuclease Activity of the Junín Virus Nucleoprotein C-Terminal Domain.Viruses. 2023 Aug 26;15(9):1818. doi: 10.3390/v15091818. Viruses. 2023. PMID: 37766225 Free PMC article.
-
Highly Pathogenic New World Arenavirus Infection Activates the Pattern Recognition Receptor Protein Kinase R without Attenuating Virus Replication in Human Cells.J Virol. 2017 Sep 27;91(20):e01090-17. doi: 10.1128/JVI.01090-17. Print 2017 Oct 15. J Virol. 2017. PMID: 28794024 Free PMC article.
-
Role of the ERK1/2 Signaling Pathway in the Replication of Junín and Tacaribe Viruses.Viruses. 2018 Apr 17;10(4):199. doi: 10.3390/v10040199. Viruses. 2018. PMID: 29673133 Free PMC article.
References
-
- Radoshitzky SR, Bao Y, Buchmeier MJ, Charrel RN, Clawson AN, Clegg CS, DeRisi JL, Emonet S, Gonzalez JP, Kuhn JH, Lukashevich IS, Peters CJ, Romanowski V, Salvato MS, Stenglein MD, de la Torre JC. 2015. Past, present, and future of arenavirus taxonomy. Arch Virol 160:1851–1874. doi:10.1007/s00705-015-2418-y. - DOI - PubMed
-
- Martínez-Peralta LA, Coto CE, Weissenbacher MC. 1993. The Tacaribe complex: the close relationship between a pathogenic (Junin) and a nonpathogenic (Tacaribe) arenavirus, p 281–296. In Salvato MS. (ed), The Arenaviridae. Plenum Press, New York, NY.
-
- Buchmeier MJ, de La Torre JC, Peters CJ. 2007. Arenaviridae: the viruses and their replication, p 1791–1828. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA.
-
- Franze-Fernández MT, Iapalucci S, López N, Rossi C. 1993. Subgenomic RNAs of Tacaribe virus, p 113–132. In Salvato MS. (ed), The Arenaviridae. Plenum Press, New York, NY.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

