Residues in the Nucleosome Acidic Patch Regulate Histone Occupancy and Are Important for FACT Binding in Saccharomyces cerevisiae
- PMID: 28468903
- PMCID: PMC5500134
- DOI: 10.1534/genetics.117.201939
Residues in the Nucleosome Acidic Patch Regulate Histone Occupancy and Are Important for FACT Binding in Saccharomyces cerevisiae
Abstract
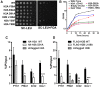
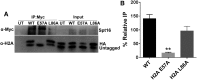
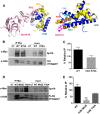
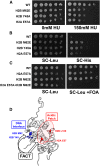
The essential histone chaperone FACT plays a critical role in DNA replication, repair, and transcription, primarily by binding to histone H2A-H2B dimers and regulating their assembly into nucleosomes. While FACT histone chaperone activity has been extensively studied, the exact nature of the H2A and H2B residues important for FACT binding remains controversial. In this study, we characterized the functions of residues in the histone H2A and H2B acidic patch, which is important for binding many chromatin-associated factors. We found that mutations in essential acidic patch residues cause a defect in histone occupancy in yeast, even though most of these histone mutants are expressed normally in yeast and form stable nucleosomes in vitro Instead, we show that two acidic patch residues, H2B L109 and H2A E57, are important for histone binding to FACT in vivo We systematically screened mutants in other H2A and H2B residues previously suspected to be important for FACT binding and confirmed the importance of H2B M62 using an in-vivo FACT-binding assay. Furthermore, we show that, like deletion mutants in FACT subunits, an H2A E57 and H2B M62 double mutant is lethal in yeast. In summary, we show that residues in the nucleosome acidic patch promote histone occupancy and are important for FACT binding to H2A-H2B dimers in yeast.
Keywords: H2A-H2B dimer; Nap1; Pob3; Spt16; nucleosome assembly.
Copyright © 2017 by the Genetics Society of America.
Figures

Similar articles
-
FACT Disrupts Nucleosome Structure by Binding H2A-H2B with Conserved Peptide Motifs.Mol Cell. 2015 Oct 15;60(2):294-306. doi: 10.1016/j.molcel.2015.09.008. Epub 2015 Oct 8. Mol Cell. 2015. PMID: 26455391 Free PMC article.
-
Structural evidence for Nap1-dependent H2A-H2B deposition and nucleosome assembly.EMBO J. 2016 Jul 1;35(13):1465-82. doi: 10.15252/embj.201694105. Epub 2016 May 25. EMBO J. 2016. PMID: 27225933 Free PMC article.
-
Insight into the mechanism of nucleosome reorganization from histone mutants that suppress defects in the FACT histone chaperone.Genetics. 2011 Aug;188(4):835-46. doi: 10.1534/genetics.111.128769. Epub 2011 May 30. Genetics. 2011. PMID: 21625001 Free PMC article.
-
Transcription through chromatin: understanding a complex FACT.Biochim Biophys Acta. 2004 Mar 15;1677(1-3):87-99. doi: 10.1016/j.bbaexp.2003.09.017. Biochim Biophys Acta. 2004. PMID: 15020050 Review.
-
[Structure and function of histone chaperone FACT].Mol Biol (Mosk). 2015 Nov-Dec;49(6):891-904. doi: 10.7868/S0026898415060026. Mol Biol (Mosk). 2015. PMID: 26710768 Review. Russian.
Cited by
-
A genetic analysis reveals novel histone residues required for transcriptional reprogramming upon stress.Nucleic Acids Res. 2020 Apr 17;48(7):3455-3475. doi: 10.1093/nar/gkaa081. Nucleic Acids Res. 2020. PMID: 32064518 Free PMC article.
-
Evidence that dissociation of Spt16 from transcribed genes is partially dependent on RNA Polymerase II termination.Transcription. 2019 Aug-Oct;10(4-5):195-206. doi: 10.1080/21541264.2019.1685837. Epub 2019 Dec 6. Transcription. 2019. PMID: 31809228 Free PMC article.
-
Essential histone chaperones collaborate to regulate transcription and chromatin integrity.Genes Dev. 2021 May 1;35(9-10):698-712. doi: 10.1101/gad.348431.121. Epub 2021 Apr 22. Genes Dev. 2021. PMID: 33888559 Free PMC article.
-
Regulation of chromatin structure and function: insights into the histone chaperone FACT.Cell Cycle. 2021 Mar-Mar;20(5-6):465-479. doi: 10.1080/15384101.2021.1881726. Epub 2021 Feb 16. Cell Cycle. 2021. PMID: 33590780 Free PMC article. Review.
-
DNA Repair in Nucleosomes: Insights from Histone Modifications and Mutants.Int J Mol Sci. 2024 Apr 16;25(8):4393. doi: 10.3390/ijms25084393. Int J Mol Sci. 2024. PMID: 38673978 Free PMC article. Review.
References
-
- Avvakumov N., Nourani A., Cote J., 2011. Histone chaperones: modulators of chromatin marks. Mol. Cell 41: 502–514. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

