A Slowed Cell Cycle Stabilizes the Budding Yeast Genome
- PMID: 28468908
- PMCID: PMC5499188
- DOI: 10.1534/genetics.116.197590
A Slowed Cell Cycle Stabilizes the Budding Yeast Genome
Abstract
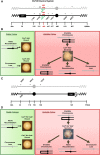
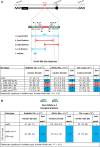
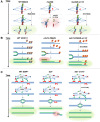
During cell division, aberrant DNA structures are detected by regulators called checkpoints that slow division to allow error correction. In addition to checkpoint-induced delay, it is widely assumed, though rarely shown, that merely slowing the cell cycle might allow more time for error detection and correction, thus resulting in a more stable genome. Fidelity by a slowed cell cycle might be independent of checkpoints. Here we tested the hypothesis that a slowed cell cycle stabilizes the genome, independent of checkpoints, in the budding yeast Saccharomyces cerevisiae We were led to this hypothesis when we identified a gene (ERV14, an ER cargo membrane protein) that when mutated, unexpectedly stabilized the genome, as measured by three different chromosome assays. After extensive studies of pathways rendered dysfunctional in erv14 mutant cells, we are led to the inference that no particular pathway is involved in stabilization, but rather the slowed cell cycle induced by erv14 stabilized the genome. We then demonstrated that, in genetic mutations and chemical treatments unrelated to ERV14, a slowed cell cycle indeed correlates with a more stable genome, even in checkpoint-proficient cells. Data suggest a delay in G2/M may commonly stabilize the genome. We conclude that chromosome errors are more rarely made or are more readily corrected when the cell cycle is slowed (even ∼15 min longer in an ∼100-min cell cycle). And, some chromosome errors may not signal checkpoint-mediated responses, or do not sufficiently signal to allow correction, and their correction benefits from this "time checkpoint."
Keywords: accuracy; chromosome instability; delayed cell cycle; erv14; speed.
Copyright © 2017 by the Genetics Society of America.
Figures



Similar articles
-
Dual cell cycle checkpoints sensitive to chromosome replication and DNA damage in the budding yeast Saccharomyces cerevisiae.Radiat Res. 1992 Nov;132(2):141-3. Radiat Res. 1992. PMID: 1438694 Review.
-
Adaptation to DNA damage checkpoint in senescent telomerase-negative cells promotes genome instability.Genes Dev. 2018 Dec 1;32(23-24):1499-1513. doi: 10.1101/gad.318485.118. Epub 2018 Nov 21. Genes Dev. 2018. PMID: 30463903 Free PMC article.
-
Ensuring the stability of the genome: DNA damage checkpoints.ScientificWorldJournal. 2001 Nov 20;1:684-702. doi: 10.1100/tsw.2001.297. ScientificWorldJournal. 2001. PMID: 12805771 Free PMC article. Review.
-
Multiple approaches to study S. cerevisiae Rad9, a prototypical checkpoint protein.Methods Enzymol. 2006;409:131-50. doi: 10.1016/S0076-6879(05)09008-7. Methods Enzymol. 2006. PMID: 16793399
-
Abrogation of the Chk1-Pds1 checkpoint leads to tolerance of persistent single-strand breaks in Saccharomyces cerevisiae.Genetics. 2005 Apr;169(4):1833-44. doi: 10.1534/genetics.104.035931. Epub 2005 Jan 31. Genetics. 2005. PMID: 15687272 Free PMC article.
Cited by
-
Repression of essential cell cycle genes increases cellular fitness.PLoS Genet. 2022 Aug 29;18(8):e1010349. doi: 10.1371/journal.pgen.1010349. eCollection 2022 Aug. PLoS Genet. 2022. PMID: 36037231 Free PMC article.
-
Budding yeast complete DNA synthesis after chromosome segregation begins.Nat Commun. 2020 May 8;11(1):2267. doi: 10.1038/s41467-020-16100-3. Nat Commun. 2020. PMID: 32385287 Free PMC article.
-
Suppression of chromosome instability by targeting a DNA helicase in budding yeast.Mol Biol Cell. 2023 Jan 1;34(1):ar3. doi: 10.1091/mbc.E22-09-0395. Epub 2022 Nov 9. Mol Biol Cell. 2023. PMID: 36350688 Free PMC article.
-
Exonuclease domain mutants of yeast DIS3 display genome instability.Nucleus. 2019 Dec;10(1):21-32. doi: 10.1080/19491034.2019.1578600. Nucleus. 2019. PMID: 30724665 Free PMC article.
-
Loss of Cdc13 causes genome instability by a deficiency in replication-dependent telomere capping.PLoS Genet. 2020 Apr 14;16(4):e1008733. doi: 10.1371/journal.pgen.1008733. eCollection 2020 Apr. PLoS Genet. 2020. PMID: 32287268 Free PMC article.
References
-
- Andersson D. I., van Verseveld H. W., Stouthamer A. H., Kurland C. G., 1986. Hicrobiology suboptimal growth with hyper-accurate ribosomes. Arch. Microbiol. 144: 96–101. - PubMed
-
- Baryshnikova A., Andrews B., 2012. Neighboring-gene effect : a genetic uncertainty principle. nature publishing group 9 (4). Nature Methods 9: 341, 343. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

