Safeguarding Our Genetic Resources with Libraries of Doubled-Haploid Lines
- PMID: 28468909
- PMCID: PMC5500154
- DOI: 10.1534/genetics.115.186205
Safeguarding Our Genetic Resources with Libraries of Doubled-Haploid Lines
Abstract
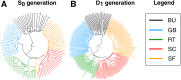
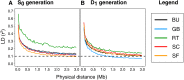
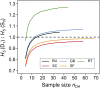
Thousands of landraces are stored in seed banks as "gold reserves" for future use in plant breeding. In many crops, their utilization is hampered because they represent heterogeneous populations of heterozygous genotypes, which harbor a high genetic load. We show, with high-density genotyping in five landraces of maize, that libraries of doubled-haploid (DH) lines capture the allelic diversity of genetic resources in an unbiased way. By comparing allelic differentiation between heterozygous plants from the original landraces and 266 derived DH lines, we find conclusive evidence that, in the DH production process, sampling of alleles is random across the entire allele frequency spectrum, and purging of landraces from their genetic load does not act on specific genomic regions. Based on overall process efficiency, we show that generating DH lines is feasible for genetic material that has never been selected for inbreeding tolerance. We conclude that libraries of DH lines will make genetic resources accessible to crop improvement by linking molecular inventories of seed banks with meaningful phenotypes.
Keywords: allelic diversity; genetic load; haploidy linkage disequilibrium; maize.
Copyright © 2017 by the Genetics Society of America.
Figures

References
-
- Aulchenko Y. S., Ripke S., Isaacs A., van Duijn C. M., 2007. GenABEL: an R library for genome-wide association analysis. Bioinformatics 23: 1294–1296. - PubMed
-
- Böhm J., Schipprack W., Mirdita V., Utz H. F., Melchinger A. E., 2014. Breeding potential of European flint maize landraces evaluated by their testcross performance. Crop Sci. 54: 1665–1672.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

