Regulation of circular dorsal ruffles, macropinocytosis, and cell migration by RhoG and its exchange factor, Trio
- PMID: 28468978
- PMCID: PMC5491185
- DOI: 10.1091/mbc.E16-06-0412
Regulation of circular dorsal ruffles, macropinocytosis, and cell migration by RhoG and its exchange factor, Trio
Abstract
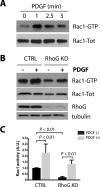
Circular dorsal ruffles (CDRs) are actin-rich structures that form on the dorsal surface of many mammalian cells in response to growth factor stimulation. CDRs represent a unique type of structure that forms transiently and only once upon stimulation. The formation of CDRs involves a drastic rearrangement of the cytoskeleton, which is regulated by the Rho family of GTPases. So far, only Rac1 has been consistently associated with CDR formation, whereas the role of other GTPases in this process is either lacking or inconclusive. Here we show that RhoG and its exchange factor, Trio, play a role in the regulation of CDR dynamics, particularly by modulating their size. RhoG is activated by Trio downstream of PDGF in a PI3K- and Src-dependent manner. Silencing RhoG expression decreases the number of cells that form CDRs, as well as the area of the CDRs. The regulation of CDR area by RhoG is independent of Rac1 function. In addition, our results show the RhoG plays a role in the cellular functions associated with CDR formation, including macropinocytosis, receptor internalization, and cell migration. Taken together, our results reveal a novel role for RhoG in the regulation of CDRs and the cellular processes associated with their formation.
© 2017 Valdivia et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures





Similar articles
-
The Rac1- and RhoG-specific GEF domain of Trio targets filamin to remodel cytoskeletal actin.Nat Cell Biol. 2000 Dec;2(12):888-92. doi: 10.1038/35046533. Nat Cell Biol. 2000. PMID: 11146652
-
Endogenous RhoG is rapidly activated after epidermal growth factor stimulation through multiple guanine-nucleotide exchange factors.Mol Biol Cell. 2010 May 1;21(9):1629-42. doi: 10.1091/mbc.e09-09-0809. Epub 2010 Mar 17. Mol Biol Cell. 2010. PMID: 20237158 Free PMC article.
-
The Rho-guanine nucleotide exchange factor Trio controls leukocyte transendothelial migration by promoting docking structure formation.Mol Biol Cell. 2012 Aug;23(15):2831-44. doi: 10.1091/mbc.E11-11-0907. Epub 2012 Jun 13. Mol Biol Cell. 2012. PMID: 22696684 Free PMC article.
-
The many faces of the guanine-nucleotide exchange factor trio.Cell Adh Migr. 2012 Nov-Dec;6(6):482-7. doi: 10.4161/cam.21418. Epub 2012 Oct 17. Cell Adh Migr. 2012. PMID: 23076143 Free PMC article. Review.
-
Roles of Rho-family GTPases in cell polarisation and directional migration.Curr Opin Cell Biol. 2003 Oct;15(5):590-7. doi: 10.1016/s0955-0674(03)00097-8. Curr Opin Cell Biol. 2003. PMID: 14519394 Review.
Cited by
-
Atypical Macropinocytosis Contributes to Malignant Progression: A Review of Recent Evidence in Endometrioid Endometrial Cancer Cells.Cancers (Basel). 2022 Oct 15;14(20):5056. doi: 10.3390/cancers14205056. Cancers (Basel). 2022. PMID: 36291839 Free PMC article. Review.
-
FARP1, ARHGEF39, and TIAM2 are essential receptor tyrosine kinase effectors for Rac1-dependent cell motility in human lung adenocarcinoma.Cell Rep. 2021 Nov 2;37(5):109905. doi: 10.1016/j.celrep.2021.109905. Cell Rep. 2021. PMID: 34731623 Free PMC article.
-
Trio family proteins as regulators of cell migration and morphogenesis in development and disease - mechanisms and cellular contexts.J Cell Sci. 2021 Feb 10;134(3):jcs248393. doi: 10.1242/jcs.248393. J Cell Sci. 2021. PMID: 33568469 Free PMC article. Review.
-
Circular dorsal ruffles disturb the growth factor-induced PI3K-AKT pathway in hepatocellular carcinoma Hep3B cells.Cell Commun Signal. 2022 Jul 7;20(1):102. doi: 10.1186/s12964-022-00911-6. Cell Commun Signal. 2022. PMID: 35799301 Free PMC article.
-
Tubeimoside 1 Acts as a Chemotherapeutic Synergist via Stimulating Macropinocytosis.Front Pharmacol. 2018 Sep 26;9:1044. doi: 10.3389/fphar.2018.01044. eCollection 2018. Front Pharmacol. 2018. PMID: 30319403 Free PMC article.
References
-
- Abercrombie M, Heaysman JE, Pegrum SM. The locomotion of fibroblasts in culture. II. “Ruffling.”. Exp Cell Res. 1970;60:437–444. - PubMed
-
- Bellanger JM, Astier C, Sardet C, Ohta Y, Stossel TP, Debant A. The Rac1- and RhoG-specific GEF domain of Trio targets filamin to remodel cytoskeletal actin. Nat Cell Biol. 2000;2:888–892. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

