Broadly neutralizing antibodies with few somatic mutations and hepatitis C virus clearance
- PMID: 28469084
- PMCID: PMC5414559
- DOI: 10.1172/jci.insight.92872
Broadly neutralizing antibodies with few somatic mutations and hepatitis C virus clearance
Abstract
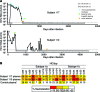
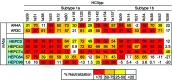
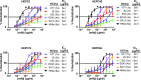
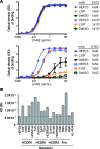
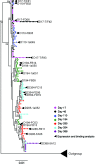
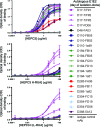
Here, we report the isolation of broadly neutralizing mAbs (bNAbs) from persons with broadly neutralizing serum who spontaneously cleared hepatitis C virus (HCV) infection. We found that bNAbs from two donors bound the same epitope and were encoded by the same germline heavy chain variable gene segment. Remarkably, these bNAbs were encoded by antibody variable genes with sparse somatic mutations. For one of the most potent bNAbs, these somatic mutations were critical for antibody neutralizing breadth and for binding to autologous envelope variants circulating late in infection. However, somatic mutations were not necessary for binding of the bNAb unmutated ancestor to envelope proteins of early autologous transmitted/founder viruses. This study identifies a public B cell clonotype favoring early recognition of a conserved HCV epitope, proving that anti-HCV bNAbs can achieve substantial neutralizing breadth with relatively few somatic mutations, and identifies HCV envelope variants that favored selection and maturation of an anti-HCV bNAb in vivo. These data provide insight into the molecular mechanisms of immune-mediated clearance of HCV infection and present a roadmap to guide development of a vaccine capable of stimulating anti-HCV bNAbs with a physiologic number of somatic mutations characteristic of vaccine responses.
Keywords: Infectious disease.
Conflict of interest statement
Figures


References
-
- Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57(4):1333–1342. - PubMed
-
- Cox AL. MEDICINE. Global control of hepatitis C virus. Science. 2015;349(6250):790–791. - PubMed
-
- Suryaprasad AG, et al. Emerging epidemic of hepatitis C virus infections among young nonurban persons who inject drugs in the United States, 2006-2012. Clin Infect Dis. 2014;59(10):1411–1419. - PubMed
Grants and funding
- UL1 TR000445/TR/NCATS NIH HHS/United States
- P30 AI094189/AI/NIAID NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- R01 AI127469/AI/NIAID NIH HHS/United States
- K08 AI102761/AI/NIAID NIH HHS/United States
- T32 AI007247/AI/NIAID NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- HHSN272201400058C/AI/NIAID NIH HHS/United States
- UL1 RR024975/RR/NCRR NIH HHS/United States
- T32 GM008752/GM/NIGMS NIH HHS/United States
- U19 AI088791/AI/NIAID NIH HHS/United States
- P30 AI045008/AI/NIAID NIH HHS/United States
- HHSN272200900055C/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

