Allosteric conformational changes of human HBV core protein transform its assembly
- PMID: 28469174
- PMCID: PMC5431180
- DOI: 10.1038/s41598-017-01568-9
Allosteric conformational changes of human HBV core protein transform its assembly
Abstract
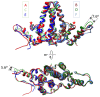
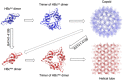
Hepatitis B Virus core protein (HBc) has multiple roles in the viral lifecycle: viral assembly, compartment for reverse transcription, intracellular trafficking, and nuclear functions. HBc displays assembly polymorphism - it can assemble into icosahedral capsid and aberrant non-capsid structures. It has been hypothesized that the assembly polymorphism is due to allosteric conformational changes of HBc dimer, the smallest assembly unit, however, the mechanism governing the polymorphic assembly of the HBc dimer is still elusive. By using the experimental antiviral drug BAY 41-4109, we successfully transformed the HBc assembly from icosahedral capsid to helical tube. Structural analyses of HBc dimers from helical tubes, T = 4 icosahedral capsid, and sheet-like HBc ensemble revealed differences within the inter-dimer interface. Disruption of the HBc inter-dimer interface may likely promote the various assembly forms of HBc. Our work provides new structural insights into the HBV assembly mechanism and strategic guide for anti-HBV drug design.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Cell-Free Hepatitis B Virus Capsid Assembly Dependent on the Core Protein C-Terminal Domain and Regulated by Phosphorylation.J Virol. 2016 May 27;90(12):5830-5844. doi: 10.1128/JVI.00394-16. Print 2016 Jun 15. J Virol. 2016. PMID: 27076641 Free PMC article.
-
Structural Differences between the Woodchuck Hepatitis Virus Core Protein in the Dimer and Capsid States Are Consistent with Entropic and Conformational Regulation of Assembly.J Virol. 2019 Jun 28;93(14):e00141-19. doi: 10.1128/JVI.00141-19. Print 2019 Jul 15. J Virol. 2019. PMID: 31043524 Free PMC article.
-
Regulation of Hepatitis B Virus Replication by Cyclin Docking Motifs in Core Protein.J Virol. 2021 May 24;95(12):e00230-21. doi: 10.1128/JVI.00230-21. Print 2021 May 24. J Virol. 2021. PMID: 33789995 Free PMC article.
-
The diverse functions of the hepatitis B core/capsid protein (HBc) in the viral life cycle: Implications for the development of HBc-targeting antivirals.Antiviral Res. 2018 Jan;149:211-220. doi: 10.1016/j.antiviral.2017.11.015. Epub 2017 Nov 26. Antiviral Res. 2018. PMID: 29183719 Free PMC article. Review.
-
Core protein: A pleiotropic keystone in the HBV lifecycle.Antiviral Res. 2015 Sep;121:82-93. doi: 10.1016/j.antiviral.2015.06.020. Epub 2015 Jun 27. Antiviral Res. 2015. PMID: 26129969 Free PMC article. Review.
Cited by
-
Hepatitis B virus core protein allosteric modulators can distort and disrupt intact capsids.Elife. 2018 Jan 29;7:e31473. doi: 10.7554/eLife.31473. Elife. 2018. PMID: 29377794 Free PMC article.
-
Hepatitis B Core Protein Capsids.Subcell Biochem. 2021;96:451-470. doi: 10.1007/978-3-030-58971-4_14. Subcell Biochem. 2021. PMID: 33252740 Review.
-
Review on hepatitis B virus precore/core promoter mutations and their correlation with genotypes and liver disease severity.World J Hepatol. 2022 Apr 27;14(4):708-718. doi: 10.4254/wjh.v14.i4.708. World J Hepatol. 2022. PMID: 35646275 Free PMC article. Review.
-
Analytical Techniques to Characterize the Structure, Properties, and Assembly of Virus Capsids.Anal Chem. 2019 Jan 2;91(1):622-636. doi: 10.1021/acs.analchem.8b04824. Epub 2018 Dec 3. Anal Chem. 2019. PMID: 30383361 Free PMC article. Review.
-
Viral structural proteins as targets for antivirals.Curr Opin Virol. 2020 Dec;45:43-50. doi: 10.1016/j.coviro.2020.07.001. Epub 2020 Aug 7. Curr Opin Virol. 2020. PMID: 32777753 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

