Antagonistic evolution of an antibiotic and its molecular chaperone: how to maintain a vital ectosymbiosis in a highly fluctuating habitat
- PMID: 28469247
- PMCID: PMC5431198
- DOI: 10.1038/s41598-017-01626-2
Antagonistic evolution of an antibiotic and its molecular chaperone: how to maintain a vital ectosymbiosis in a highly fluctuating habitat
Erratum in
-
Author Correction: Antagonistic evolution of an antibiotic and its molecular chaperone: how to maintain a vital ectosymbiosis in a highly fluctuating habitat.Sci Rep. 2017 Dec 4;7(1):17154. doi: 10.1038/s41598-017-16211-w. Sci Rep. 2017. PMID: 29203774 Free PMC article.
Abstract
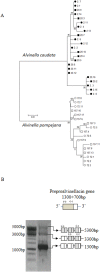
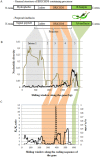
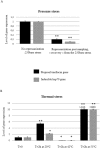
Evolution of antimicrobial peptides (AMPs) has been shown to be driven by recurrent duplications and balancing/positive selection in response to new or altered bacterial pathogens. We use Alvinella pompejana, the most eurythermal animal known on Earth, to decipher the selection patterns acting on AMP in an ecological rather than controlled infection approach. The preproalvinellacin multigenic family presents the uniqueness to encode a molecular chaperone (BRICHOS) together with an AMP (alvinellacin) that controls the vital ectosymbiosis of Alvinella. In stark contrast to what is observed in the context of the Red queen paradigm, we demonstrate that exhibiting a vital and highly conserved ecto-symbiosis in the face of thermal fluctuations has led to a peculiar selective trend promoting the adaptive diversification of the molecular chaperone of the AMP, but not of the AMP itself. Because BRICHOS stabilizes beta-stranded peptides, this polymorphism likely represents an eurythermal adaptation to stabilize the structure of alvinellacin, thus hinting at its efficiency to select and control the epibiosis across the range of temperatures experienced by the worm; Our results fill some knowledge gaps concerning the function of BRICHOS in invertebrates and offer perspectives for studying immune genes in an evolutionary ecological framework.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

