Automated multiplex genome-scale engineering in yeast
- PMID: 28469255
- PMCID: PMC5418614
- DOI: 10.1038/ncomms15187
Automated multiplex genome-scale engineering in yeast
Abstract
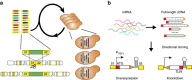
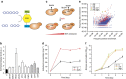
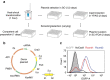
Genome-scale engineering is indispensable in understanding and engineering microorganisms, but the current tools are mainly limited to bacterial systems. Here we report an automated platform for multiplex genome-scale engineering in Saccharomyces cerevisiae, an important eukaryotic model and widely used microbial cell factory. Standardized genetic parts encoding overexpression and knockdown mutations of >90% yeast genes are created in a single step from a full-length cDNA library. With the aid of CRISPR-Cas, these genetic parts are iteratively integrated into the repetitive genomic sequences in a modular manner using robotic automation. This system allows functional mapping and multiplex optimization on a genome scale for diverse phenotypes including cellulase expression, isobutanol production, glycerol utilization and acetic acid tolerance, and may greatly accelerate future genome-scale engineering endeavours in yeast.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Carr P. A. & Church G. M. Genome engineering. Nat. Biotechnol. 27, 1151–1162 (2009). - PubMed
-
- Giaever G. et al.. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391 (2002). - PubMed
-
- Warner J. R., Reeder P. J., Karimpour-Fard A., Woodruff L. B. & Gill R. T. Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides. Nat. Biotechnol. 28, 856–862 (2010). - PubMed
-
- Si T., Luo Y., Bao Z. & Zhao H. RNAi-assisted genome evolution in Saccharomyces cerevisiae for complex phenotype engineering. ACS Synth. Biol. 4, 283–291 (2015). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

