CRISPR-Cpf1 assisted genome editing of Corynebacterium glutamicum
- PMID: 28469274
- PMCID: PMC5418603
- DOI: 10.1038/ncomms15179
CRISPR-Cpf1 assisted genome editing of Corynebacterium glutamicum
Abstract
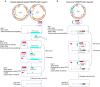
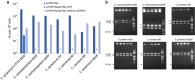
Corynebacterium glutamicum is an important industrial metabolite producer that is difficult to genetically engineer. Although the Streptococcus pyogenes (Sp) CRISPR-Cas9 system has been adapted for genome editing of multiple bacteria, it cannot be introduced into C. glutamicum. Here we report a Francisella novicida (Fn) CRISPR-Cpf1-based genome-editing method for C. glutamicum. CRISPR-Cpf1, combined with single-stranded DNA (ssDNA) recombineering, precisely introduces small changes into the bacterial genome at efficiencies of 86-100%. Large gene deletions and insertions are also obtained using an all-in-one plasmid consisting of FnCpf1, CRISPR RNA, and homologous arms. The two CRISPR-Cpf1-assisted systems enable N iterative rounds of genome editing in 3N+4 or 3N+2 days. A proof-of-concept, codon saturation mutagenesis at G149 of γ-glutamyl kinase relieves L-proline inhibition using Cpf1-assisted ssDNA recombineering. Thus, CRISPR-Cpf1-based genome editing provides a highly efficient tool for genetic engineering of Corynebacterium and other bacteria that cannot utilize the Sp CRISPR-Cas9 system.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



Similar articles
-
CRISPR-Cpf1-Assisted Multiplex Genome Editing and Transcriptional Repression in Streptomyces.Appl Environ Microbiol. 2018 Aug 31;84(18):e00827-18. doi: 10.1128/AEM.00827-18. Print 2018 Sep 15. Appl Environ Microbiol. 2018. PMID: 29980561 Free PMC article.
-
Development of a CRISPR/Cas9 genome editing toolbox for Corynebacterium glutamicum.Microb Cell Fact. 2017 Nov 16;16(1):205. doi: 10.1186/s12934-017-0815-5. Microb Cell Fact. 2017. PMID: 29145843 Free PMC article.
-
Genome Editing of Corynebacterium glutamicum Using CRISPR-Cpf1 System.Methods Mol Biol. 2022;2479:189-206. doi: 10.1007/978-1-0716-2233-9_13. Methods Mol Biol. 2022. PMID: 35583740
-
Cas9, Cpf1 and C2c1/2/3-What's next?Bioengineered. 2017 May 4;8(3):265-273. doi: 10.1080/21655979.2017.1282018. Epub 2017 Jan 31. Bioengineered. 2017. PMID: 28140746 Free PMC article. Review.
-
[The new generation tool for CRISPR genome editing: CRISPR/Cpf1].Sheng Wu Gong Cheng Xue Bao. 2017 Mar 25;33(3):361-371. doi: 10.13345/j.cjb.170029. Sheng Wu Gong Cheng Xue Bao. 2017. PMID: 28941336 Review. Chinese.
Cited by
-
High-Fidelity Cytosine Base Editing in a GC-Rich Corynebacterium glutamicum with Reduced DNA Off-Target Editing Effects.Microbiol Spectr. 2022 Dec 21;10(6):e0376022. doi: 10.1128/spectrum.03760-22. Epub 2022 Nov 14. Microbiol Spectr. 2022. PMID: 36374037 Free PMC article.
-
Establishment of the CRISPR-Cpf1 gene editing system in Bacillus licheniformis and multiplexed gene knockout.Synth Syst Biotechnol. 2024 Aug 8;10(1):39-48. doi: 10.1016/j.synbio.2024.08.002. eCollection 2025. Synth Syst Biotechnol. 2024. PMID: 39224148 Free PMC article.
-
Triple deletion of clpC, porB, and mepA enhances production of small ubiquitin-like modifier-N-terminal pro-brain natriuretic peptide in Corynebacterium glutamicum.J Ind Microbiol Biotechnol. 2019 Jan;46(1):67-79. doi: 10.1007/s10295-018-2091-8. Epub 2018 Oct 24. J Ind Microbiol Biotechnol. 2019. PMID: 30357503
-
Genome editing of lactic acid bacteria: opportunities for food, feed, pharma and biotech.FEMS Microbiol Lett. 2019 Jan 1;366(1):fny291. doi: 10.1093/femsle/fny291. FEMS Microbiol Lett. 2019. PMID: 30561594 Free PMC article. Review.
-
Use of CRISPR-Cas tools to engineer Trichoderma species.Microb Biotechnol. 2022 Oct;15(10):2521-2532. doi: 10.1111/1751-7915.14126. Epub 2022 Jul 31. Microb Biotechnol. 2022. PMID: 35908288 Free PMC article. Review.
References
-
- Wendisch V. F., Jorge J. M., Perez-Garcia F. & Sgobba E. Updates on industrial production of amino acids using Corynebacterium glutamicum. World. J. Microbiol. Biotechnol. 32, 105 (2016). - PubMed
-
- Lee J. Y., Na Y. A., Kim E., Lee H. S. & Kim P. The Actinobacterium Corynebacterium glutamicum, an Industrial Workhorse. J. Microbiol. Biotechnol. 26, 807–822 (2016). - PubMed
-
- Eggeling L. & Bott M. A giant market and a powerful metabolism: L-lysine provided by Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 99, 3387–3394 (2015). - PubMed
-
- Heider S. A. & Wendisch V. F. Engineering microbial cell factories: Metabolic engineering of Corynebacterium glutamicum with a focus on non-natural products. Biotechnol. J. 10, 1170–1184 (2015). - PubMed
-
- Nesvera J. & Patek M. Tools for genetic manipulations in Corynebacterium glutamicum and their applications. Appl. Microbiol. Biotechnol. 90, 1641–1654 (2011). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

