A mixture copula Bayesian network model for multimodal genomic data
- PMID: 28469391
- PMCID: PMC5397279
- DOI: 10.1177/1176935117702389
A mixture copula Bayesian network model for multimodal genomic data
Abstract
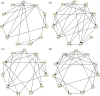
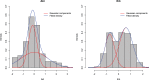
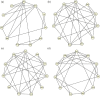
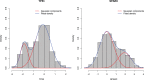
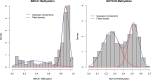
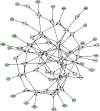
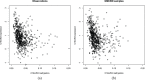
Gaussian Bayesian networks have become a widely used framework to estimate directed associations between joint Gaussian variables, where the network structure encodes the decomposition of multivariate normal density into local terms. However, the resulting estimates can be inaccurate when the normality assumption is moderately or severely violated, making it unsuitable for dealing with recent genomic data such as the Cancer Genome Atlas data. In the present paper, we propose a mixture copula Bayesian network model which provides great flexibility in modeling non-Gaussian and multimodal data for causal inference. The parameters in mixture copula functions can be efficiently estimated by a routine expectation-maximization algorithm. A heuristic search algorithm based on Bayesian information criterion is developed to estimate the network structure, and prediction can be further improved by the best-scoring network out of multiple predictions from random initial values. Our method outperforms Gaussian Bayesian networks and regular copula Bayesian networks in terms of modeling flexibility and prediction accuracy, as demonstrated using a cell signaling data set. We apply the proposed methods to the Cancer Genome Atlas data to study the genetic and epigenetic pathways that underlie serous ovarian cancer.
Keywords: Bayesian network; copula function; serous ovarian cancer; systems biology; the Cancer Genome Atlas.
Conflict of interest statement
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures

Similar articles
-
A new mixture copula model for spatially correlated multiple variables with an environmental application.Sci Rep. 2022 Aug 16;12(1):13867. doi: 10.1038/s41598-022-18007-z. Sci Rep. 2022. PMID: 35974067 Free PMC article.
-
Constructing gene regulatory networks from microarray data using non-Gaussian pair-copula Bayesian networks.J Bioinform Comput Biol. 2020 Aug;18(4):2050023. doi: 10.1142/S0219720020500237. Epub 2020 Jul 24. J Bioinform Comput Biol. 2020. PMID: 32706288
-
A two-level copula joint model for joint analysis of longitudinal and competing risks data.Stat Med. 2023 May 30;42(12):1909-1930. doi: 10.1002/sim.9704. Epub 2023 Mar 7. Stat Med. 2023. PMID: 37194500
-
A spatial copula interpolation in a random field with application in air pollution data.Model Earth Syst Environ. 2023;9(1):175-194. doi: 10.1007/s40808-022-01484-6. Epub 2022 Aug 18. Model Earth Syst Environ. 2023. PMID: 35996594 Free PMC article.
-
IDENTIFYING THE NUMBER OF COMPONENTS IN GAUSSIAN MIXTURE MODELS USING NUMERICAL ALGEBRAIC GEOMETRY.J Algebra Appl. 2020 Nov;19(11):2050204. doi: 10.1142/s0219498820502047. Epub 2019 Oct 21. J Algebra Appl. 2020. PMID: 33867617 Free PMC article.
Cited by
-
A new mixture copula model for spatially correlated multiple variables with an environmental application.Sci Rep. 2022 Aug 16;12(1):13867. doi: 10.1038/s41598-022-18007-z. Sci Rep. 2022. PMID: 35974067 Free PMC article.
-
Synthetic data generation with probabilistic Bayesian Networks.Math Biosci Eng. 2021 Oct 9;18(6):8603-8621. doi: 10.3934/mbe.2021426. Math Biosci Eng. 2021. PMID: 34814315 Free PMC article.
References
-
- Fu F, Zhou Q. Learning sparse causal Gaussian networks with experimental intervention: Regularization and coordinate descent. J Amer Stat Assoc. 2013;108(501):288–300.
-
- Friedman N, Linial M, Nachman I, et al. Using Bayesian networks to analyze expression data. J Computat Biol. 2000;7(3):601–20. - PubMed
-
- Ellis B, Wong WH. Learning causal Bayesian network structures from experimental data. J Amer Stat Assoc. 2008;103(482):778–789.
LinkOut - more resources
Full Text Sources
Other Literature Sources

