Evolution of Dimethylsulfoniopropionate Metabolism in Marine Phytoplankton and Bacteria
- PMID: 28469605
- PMCID: PMC5395565
- DOI: 10.3389/fmicb.2017.00637
Evolution of Dimethylsulfoniopropionate Metabolism in Marine Phytoplankton and Bacteria
Abstract
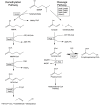
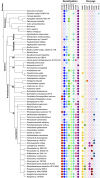
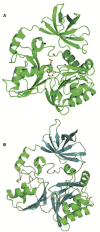
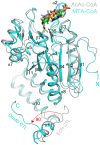
The elucidation of the pathways for dimethylsulfoniopropionate (DMSP) synthesis and metabolism and the ecological impact of DMSP have been studied for nearly 70 years. Much of this interest stems from the fact that DMSP metabolism produces the climatically active gas dimethyl sulfide (DMS), the primary natural source of sulfur to the atmosphere. DMSP plays many important roles for marine life, including use as an osmolyte, antioxidant, predator deterrent, and cryoprotectant for phytoplankton and as a reduced carbon and sulfur source for marine bacteria. DMSP is hypothesized to have become abundant in oceans approximately 250 million years ago with the diversification of the strong DMSP producers, the dinoflagellates. This event coincides with the first genome expansion of the Roseobacter clade, known DMSP degraders. Structural and mechanistic studies of the enzymes of the bacterial DMSP demethylation and cleavage pathways suggest that exposure to DMSP led to the recruitment of enzymes from preexisting metabolic pathways. In some cases, such as DmdA, DmdD, and DddP, these enzymes appear to have evolved to become more specific for DMSP metabolism. By contrast, many of the other enzymes, DmdB, DmdC, and the acrylate utilization hydratase AcuH, have maintained broad functionality and substrate specificities, allowing them to carry out a range of reactions within the cell. This review will cover the experimental evidence supporting the hypothesis that, as DMSP became more readily available in the marine environment, marine bacteria adapted enzymes already encoded in their genomes to utilize this new compound.
Keywords: DMSP; Roseobacter; dimethylsulfoniopropionate; evolution; phytoplankton.
Figures

Similar articles
-
Mechanistic insight into 3-methylmercaptopropionate metabolism and kinetical regulation of demethylation pathway in marine dimethylsulfoniopropionate-catabolizing bacteria.Mol Microbiol. 2019 Apr;111(4):1057-1073. doi: 10.1111/mmi.14211. Epub 2019 Mar 4. Mol Microbiol. 2019. PMID: 30677184 Free PMC article.
-
Bacterial Catabolism of Dimethylsulfoniopropionate (DMSP).Front Microbiol. 2011 Aug 12;2:172. doi: 10.3389/fmicb.2011.00172. eCollection 2011. Front Microbiol. 2011. PMID: 21886640 Free PMC article.
-
Recent insights into oceanic dimethylsulfoniopropionate biosynthesis and catabolism.Environ Microbiol. 2022 Jun;24(6):2669-2700. doi: 10.1111/1462-2920.16045. Epub 2022 May 25. Environ Microbiol. 2022. PMID: 35611751 Review.
-
Biogenic production of DMSP and its degradation to DMS-their roles in the global sulfur cycle.Sci China Life Sci. 2019 Oct;62(10):1296-1319. doi: 10.1007/s11427-018-9524-y. Epub 2019 Jun 20. Sci China Life Sci. 2019. PMID: 31231779 Review.
-
Evolutionary history of dimethylsulfoniopropionate (DMSP) demethylation enzyme DmdA in marine bacteria.PeerJ. 2020 Sep 10;8:e9861. doi: 10.7717/peerj.9861. eCollection 2020. PeerJ. 2020. PMID: 32974097 Free PMC article.
Cited by
-
A comparative whole-genome approach identifies bacterial traits for marine microbial interactions.Commun Biol. 2022 Mar 28;5(1):276. doi: 10.1038/s42003-022-03184-4. Commun Biol. 2022. PMID: 35347228 Free PMC article.
-
Molecular Insight into the Acryloyl-CoA Hydration by AcuH for Acrylate Detoxification in Dimethylsulfoniopropionate-Catabolizing Bacteria.Front Microbiol. 2017 Oct 17;8:2034. doi: 10.3389/fmicb.2017.02034. eCollection 2017. Front Microbiol. 2017. PMID: 29089943 Free PMC article.
-
Mechanistic insight into 3-methylmercaptopropionate metabolism and kinetical regulation of demethylation pathway in marine dimethylsulfoniopropionate-catabolizing bacteria.Mol Microbiol. 2019 Apr;111(4):1057-1073. doi: 10.1111/mmi.14211. Epub 2019 Mar 4. Mol Microbiol. 2019. PMID: 30677184 Free PMC article.
-
Acrylate protects a marine bacterium from grazing by a ciliate predator.Nat Microbiol. 2021 Nov;6(11):1351-1356. doi: 10.1038/s41564-021-00981-1. Epub 2021 Oct 25. Nat Microbiol. 2021. PMID: 34697458
-
Quorum Sensing Regulates the Production of Methanethiol in Vibrio harveyi.Microorganisms. 2023 Dec 24;12(1):35. doi: 10.3390/microorganisms12010035. Microorganisms. 2023. PMID: 38257862 Free PMC article.
References
-
- Allen A. E. (2005). Defining the moelcular basis for energy balance in marine diatoms under fluctuating environmental conditions. J. Phycol. 6 1073–1076. 10.1111/j.1529-8817.2005.00156.x - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

