Integrated data analysis reveals potential drivers and pathways disrupted by DNA methylation in papillary thyroid carcinomas
- PMID: 28469731
- PMCID: PMC5414166
- DOI: 10.1186/s13148-017-0346-2
Integrated data analysis reveals potential drivers and pathways disrupted by DNA methylation in papillary thyroid carcinomas
Abstract
Background: Papillary thyroid carcinoma (PTC) is a common endocrine neoplasm with a recent increase in incidence in many countries. Although PTC has been explored by gene expression and DNA methylation studies, the regulatory mechanisms of the methylation on the gene expression was poorly clarified. In this study, DNA methylation profile (Illumina HumanMethylation 450K) of 41 PTC paired with non-neoplastic adjacent tissues (NT) was carried out to identify and contribute to the elucidation of the role of novel genic and intergenic regions beyond those described in the promoter and CpG islands (CGI). An integrative and cross-validation analysis were performed aiming to identify molecular drivers and pathways that are PTC-related.
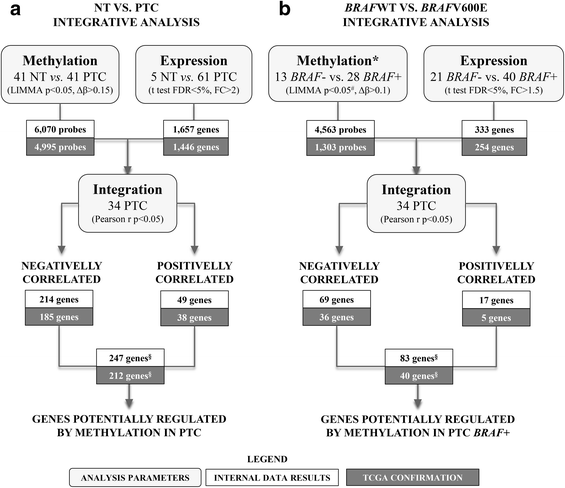
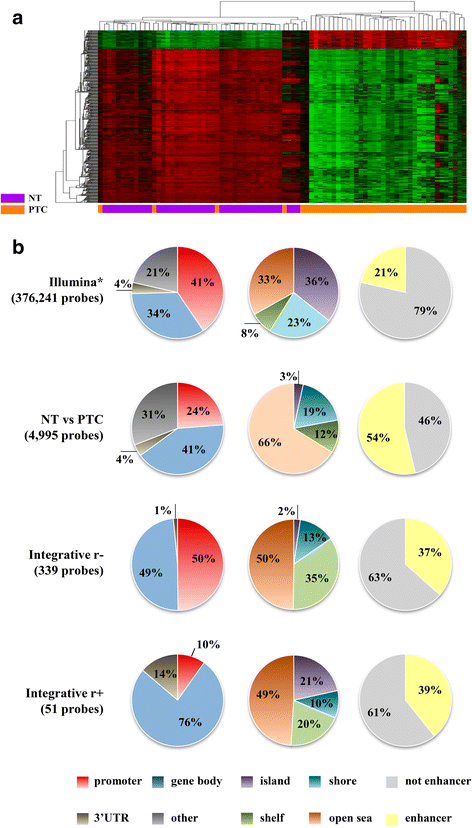
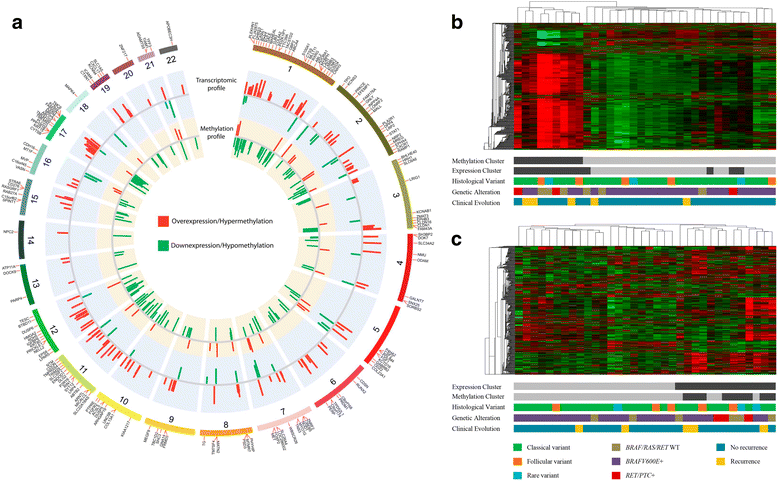
Results: The comparisons between PTC and NT revealed 4995 methylated probes (88% hypomethylated in PTC) and 1446 differentially expressed transcripts cross-validated by the The Cancer Genome Atlas data. The majority of these probes was found in non-promoters regions, distant from CGI and enriched by enhancers. The integrative analysis between gene expression and DNA methylation revealed 185 and 38 genes (mainly in the promoter and body regions, respectively) with negative and positive correlation, respectively. Genes showing negative correlation underlined FGF and retinoic acid signaling as critical canonical pathways disrupted by DNA methylation in PTC. BRAF mutation was detected in 68% (28 of 41) of the tumors, which presented a higher level of demethylation (95% hypomethylated probes) compared with BRAF wild-type tumors. A similar integrative analysis uncovered 40 of 254 differentially expressed genes, which are potentially regulated by DNA methylation in BRAFV600E-positive tumors. The methylation and expression pattern of six selected genes (ERBB3, FGF1, FGFR2, GABRB2, HMGA2, and RDH5) were confirmed as altered by pyrosequencing and RT-qPCR.
Conclusions: DNA methylation loss in non-promoter, poor CGI and enhancer-enriched regions was a significant event in PTC, especially in tumors harboring BRAFV600E. In addition to the promoter region, gene body and 3'UTR methylation have also the potential to influence the gene expression levels (both, repressing and inducing). The integrative analysis revealed genes potentially regulated by DNA methylation pointing out potential drivers and biomarkers related to PTC development.
Keywords: BRAFV600E mutation; DNA methylation; FGF signaling pathway; Integrative analysis; Papillary thyroid cancer; Retinoic acid pathway.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

