Melatonin treatment enhances therapeutic effects of exosomes against acute liver ischemia-reperfusion injury
- PMID: 28469765
- PMCID: PMC5411908
Melatonin treatment enhances therapeutic effects of exosomes against acute liver ischemia-reperfusion injury
Abstract
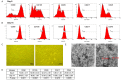
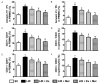
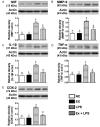
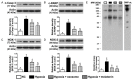
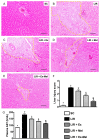
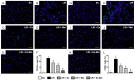
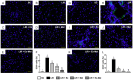
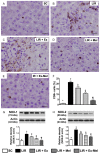
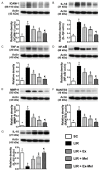
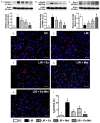
This study tests the hypothesis that combined melatonin and exogenic adipose mesenchymal stem cell (ADMSC)-derived exosome treatment offers superior protection against liver ischemia-reperfusion (LIR) injury compared to either alone. In vitro studies utilized a macrophage cell line (RAW) pretreated with lipopolysaccharide and hepatocytes pretreated with melatonin or exosomes before hypoxia treatment, while in vitro experiments involved analyses of liver specimens from male adult Sprague-Dawley rats (n = 50) equally categorized into sham controls (SC), LIR only, LIR-exosome (100 µg, 30 minute post-LIR), LIR-melatonin (20 mg/kg, 30 minute post-LIR and 50 mg/kg at 6 and 18 hours post-LIR), and LIR-exosome-melatonin groups. In vitro studies showed suppression of inflammation (MIF, MMP-9, IL-1β, TNF-α, COX-2) and oxidative stress (NOX-1, NOX-2, oxidized protein)/apoptosis (cleaved caspase 3 and PARP) by exosome and exosome/melatonin treatment, respectively (all P<0.001). In vivo data demonstrated lowest liver injury score and plasma AST concentrations in LIR-exosome-melatonin group compared with other groups (P<0.001). Besides, expressions of inflammatory markers at protein (ICAM-1, IL-1β, MMP-9, TNF-α, NF-κB, RANTES) and cellular (CD3+, CD4+, CD8+, CD161+, CD11+, CD14+, F4/80) levels, and protein expressions of apoptosis (cleaved caspase-3, PARP), oxidative stress (NOX-1, NOX-2), DNA damage (γ-H2AX) and mitochondrial damage (cytosolic cytochrome-C) markers displayed a pattern similar to that of liver injury score, whereas protein expression of anti-oxidants (HO-1, NQO-1) showed progressive increase from SC to the combined treatment group (all P<0.001). In conclusion, combined exosome-melatonin regimen was superior to either alone in protecting the liver against ischemia-reperfusion injury.
Keywords: Exogenic exosomes; anti-oxidant; immunomodulation; inflammation; liver ischemia-reperfusion injury; melatonin; oxidative stress.
Conflict of interest statement
None.
Figures



Similar articles
-
Melatonin pretreatment enhances the therapeutic effects of exogenous mitochondria against hepatic ischemia-reperfusion injury in rats through suppression of mitochondrial permeability transition.J Pineal Res. 2016 Aug;61(1):52-68. doi: 10.1111/jpi.12326. Epub 2016 Apr 7. J Pineal Res. 2016. PMID: 26993080
-
Protective effect of melatonin-supported adipose-derived mesenchymal stem cells against small bowel ischemia-reperfusion injury in rat.J Pineal Res. 2015 Sep;59(2):206-20. doi: 10.1111/jpi.12251. Epub 2015 Jun 24. J Pineal Res. 2015. PMID: 26013733
-
Combination of adipose-derived mesenchymal stem cells (ADMSC) and ADMSC-derived exosomes for protecting kidney from acute ischemia-reperfusion injury.Int J Cardiol. 2016 Aug 1;216:173-85. doi: 10.1016/j.ijcard.2016.04.061. Epub 2016 Apr 14. Int J Cardiol. 2016. PMID: 27156061
-
Sitagliptin protects rat kidneys from acute ischemia-reperfusion injury via upregulation of GLP-1 and GLP-1 receptors.Acta Pharmacol Sin. 2015 Jan;36(1):119-30. doi: 10.1038/aps.2014.98. Epub 2014 Dec 15. Acta Pharmacol Sin. 2015. PMID: 25500876 Free PMC article. Review.
-
Exosomes and Melatonin: Where Their Destinies Intersect.Front Immunol. 2021 Jun 11;12:692022. doi: 10.3389/fimmu.2021.692022. eCollection 2021. Front Immunol. 2021. PMID: 34177952 Free PMC article. Review.
Cited by
-
Melatonin pretreatment on exosomes: Heterogeneity, therapeutic effects, and usage.Front Immunol. 2022 Sep 16;13:933736. doi: 10.3389/fimmu.2022.933736. eCollection 2022. Front Immunol. 2022. PMID: 36189281 Free PMC article. Review.
-
Tailored Extracellular Vesicles: Novel Tool for Tissue Regeneration.Stem Cells Int. 2022 Jul 29;2022:7695078. doi: 10.1155/2022/7695078. eCollection 2022. Stem Cells Int. 2022. PMID: 35915850 Free PMC article. Review.
-
Extracellular Vesicles in Liver Transplantation: Current Evidence and Future Challenges.Int J Mol Sci. 2023 Aug 31;24(17):13547. doi: 10.3390/ijms241713547. Int J Mol Sci. 2023. PMID: 37686354 Free PMC article. Review.
-
An Examination of the Putative Role of Melatonin in Exosome Biogenesis.Front Cell Dev Biol. 2021 Jun 8;9:686551. doi: 10.3389/fcell.2021.686551. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34169078 Free PMC article. Review.
-
Combined tacrolimus and melatonin effectively protected kidney against acute ischemia-reperfusion injury.FASEB J. 2021 Jun;35(6):e21661. doi: 10.1096/fj.202100174R. FASEB J. 2021. PMID: 34029398 Free PMC article.
References
-
- Khashab M, Tector AJ, Kwo PY. Epidemiology of acute liver failure. Curr Gastroenterol Rep. 2007;9:66–73. - PubMed
-
- Henrion J. Hypoxic hepatitis. Liver Int. 2012;32:1039–1052. - PubMed
-
- Fuhrmann V, Kneidinger N, Herkner H, Heinz G, Nikfardjam M, Bojic A, Schellongowski P, Angermayr B, Kitzberger R, Warszawska J, Holzinger U, Schenk P, Madl C. Hypoxic hepatitis: underlying conditions and risk factors for mortality in critically ill patients. Intensive Care Med. 2009;35:1397–1405. - PubMed
-
- Lee WM. Acute liver failure. Semin Respir Crit Care Med. 2012;33:36–45. - PubMed
-
- Singanayagam A, Bernal W. Update on acute liver failure. Curr Opin Crit Care. 2015;21:134–141. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
