Mechanical Deformation Accelerates Protein Ageing
- PMID: 28470663
- PMCID: PMC5540753
- DOI: 10.1002/anie.201703630
Mechanical Deformation Accelerates Protein Ageing
Abstract
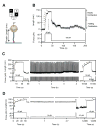
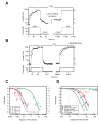
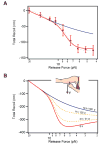
A hallmark of tissue ageing is the irreversible oxidative modification of its proteins. We show that single proteins, kept unfolded and extended by a mechanical force, undergo accelerated ageing in times scales of minutes to days. A protein forced to be continuously unfolded completely loses its ability to contract by folding, becoming a labile polymer. Ageing rates vary among different proteins, but in all cases they lose their mechanical integrity. Random oxidative modification of cryptic side chains exposed by mechanical unfolding can be slowed by the addition of antioxidants such as ascorbic acid, or accelerated by oxidants. By contrast, proteins kept in the folded state and probed over week-long experiments show greatly reduced rates of ageing. We demonstrate a novel approach whereby protein ageing can be greatly accelerated: the constant unfolding of a protein for hours to days is equivalent to decades of exposure to free radicals under physiological conditions.
Keywords: force spectroscopy; oxidative damage; protein folding; protein structure; single-molecule studies.
© 2017 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures

References
-
- Yu B, Kang SY, Akthakul A, Ramadurai N, Pilkenton M, Patel A, Nashat A, Anderson DG, Sakamoto FH, Gilchrest BA, Anderson RR, Langer R. Nature materials. 2016;15:911–918. - PubMed
-
- Nystrom T. The EMBO journal. 2005;24:1311–1317. - PMC - PubMed
- Stadtman ER. Science. 1992;257:1220–1224. - PubMed
- Gorisse L, Pietrement C, Vuiblet V, Schmelzer CE, Kohler M, Duca L, Debelle L, Fornes P, Jaisson S, Gillery P. Proc Natl Acad Sci U S A. 2016;113:1191–1196. - PMC - PubMed
- Rao RS, Moller IM. Proteomics. 2011;11:4166–4173. - PubMed
-
- Berlett BS, Stadtman ER. J Biol Chem. 1997;272:20313–20316. - PubMed
- Halliwell B, Gutteridge JMC. Free radicals in biology and medicine. Fifth. Oxford University Press; Oxford, United Kingdom: 2015.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

