Carbohydrate recognition by the rhamnose-binding lectin SUL-I with a novel three-domain structure isolated from the venom of globiferous pedicellariae of the flower sea urchin Toxopneustes pileolus
- PMID: 28470711
- PMCID: PMC5521583
- DOI: 10.1002/pro.3185
Carbohydrate recognition by the rhamnose-binding lectin SUL-I with a novel three-domain structure isolated from the venom of globiferous pedicellariae of the flower sea urchin Toxopneustes pileolus
Abstract
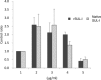
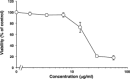
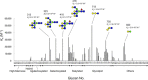
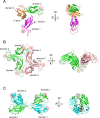
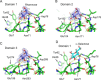
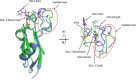
The globiferous pedicellariae of the venomous sea urchin Toxopneustes pileolus contains several biologically active proteins. We have cloned the cDNA of one of the toxin components, SUL-I, which is a rhamnose-binding lectin (RBL) that acts as a mitogen through binding to carbohydrate chains on target cells. Recombinant SUL-I (rSUL-I) was produced in Escherichia coli cells, and its carbohydrate-binding specificity was examined with the glycoconjugate microarray analysis, which suggested that potential target carbohydrate structures are galactose-terminated N-glycans. rSUL-I exhibited mitogenic activity for murine splenocyte cells and toxicity against Vero cells. The three-dimensional structure of the rSUL-I/l-rhamnose complex was determined by X-ray crystallographic analysis at a 1.8 Å resolution. The overall structure of rSUL-I is composed of three distinctive domains with a folding structure similar to those of CSL3, a RBL from chum salmon (Oncorhynchus keta) eggs. The bound l-rhamnose molecules are mainly recognized by rSUL-I through hydrogen bonds between its 2-, 3-, and 4-hydroxy groups and Asp, Asn, and Glu residues in the binding sites, while Tyr and Ser residues participate in the recognition mechanism. It was also inferred that SUL-I may form a dimer in solution based on the molecular size estimated via dynamic light scattering as well as possible contact regions in its crystal structure.
Keywords: X-ray crystallographic analysis; lectin; rhamnose; sea urchin; toxin.
© 2017 The Protein Society.
Figures




Similar articles
-
cDNA cloning and characterization of a rhamnose-binding lectin SUL-I from the toxopneustid sea urchin Toxopneustes pileolus venom.Toxicon. 2015 Feb;94:8-15. doi: 10.1016/j.toxicon.2014.11.236. Epub 2014 Dec 2. Toxicon. 2015. PMID: 25475394
-
cDNA cloning and expression of Contractin A, a phospholipase A2-like protein from the globiferous pedicellariae of the venomous sea urchin Toxopneustes pileolus.Toxicon. 2015 Dec 15;108:46-52. doi: 10.1016/j.toxicon.2015.09.040. Epub 2015 Oct 3. Toxicon. 2015. PMID: 26435342
-
Isolation of a novel lectin from the globiferous pedicellariae of the sea urchin Toxopneustes pileolus.Adv Exp Med Biol. 1996;391:213-23. doi: 10.1007/978-1-4613-0361-9_14. Adv Exp Med Biol. 1996. PMID: 8726059
-
SUEL-related lectins, a lectin family widely distributed throughout organisms.Biosci Biotechnol Biochem. 2010;74(6):1141-4. doi: 10.1271/bbb.100086. Epub 2010 Jun 7. Biosci Biotechnol Biochem. 2010. PMID: 20530910 Review.
-
The structure of sulfated polysaccharides ensures a carbohydrate-based mechanism for species recognition during sea urchin fertilization.Int J Dev Biol. 2008;52(5-6):551-9. doi: 10.1387/ijdb.072531av. Int J Dev Biol. 2008. PMID: 18649269 Review.
Cited by
-
Venom Diversity and Evolution in the Most Divergent Cone Snail Genus Profundiconus.Toxins (Basel). 2019 Oct 28;11(11):623. doi: 10.3390/toxins11110623. Toxins (Basel). 2019. PMID: 31661832 Free PMC article.
-
Evolution, Expression Patterns, and Distribution of Novel Ribbon Worm Predatory and Defensive Toxins.Mol Biol Evol. 2022 May 3;39(5):msac096. doi: 10.1093/molbev/msac096. Mol Biol Evol. 2022. PMID: 35512366 Free PMC article.
-
Functional Diversity of Novel Lectins with Unique Structural Features in Marine Animals.Cells. 2023 Jul 9;12(14):1814. doi: 10.3390/cells12141814. Cells. 2023. PMID: 37508479 Free PMC article. Review.
-
New Data on the Rhamnose-Binding Lectin from the Colonial Ascidian Botryllus schlosseri: Subcellular Distribution, Secretion Mode and Effects on the Cyclical Generation Change.Mar Drugs. 2023 Mar 8;21(3):171. doi: 10.3390/md21030171. Mar Drugs. 2023. PMID: 36976220 Free PMC article.
-
Multifunctional Cell Regulation Activities of the Mussel Lectin SeviL: Induction of Macrophage Polarization toward the M1 Functional Phenotype.Mar Drugs. 2024 Jun 11;22(6):269. doi: 10.3390/md22060269. Mar Drugs. 2024. PMID: 38921580 Free PMC article.
References
-
- Nakagawa H, Tanigawa T, Tomita K, Tomihara Y, Araki Y, Tachikawa E (2003) Recent studies on the phathological effects of purified sea urchin toxins. J Toxicol 22:633–649.
-
- Takei M, Nakagawa H (2006) A sea urchin lectin, SUL‐1, from the toxopneustid sea urchin induces DC maturation from human monocyte and drives Th1 polirization in vitro. Toxicol Appl Pharmacol 213:27–36. - PubMed
-
- Edo K, Sakai H, Nakagawa H, Hashimoto T, Shinohara M, Ohura K (2012) Immunomodulatory activity of a pedicellarial venom lectin from the toxopneustid sea urchin, Toxopneustes pileolus . Toxin Rev 31:54–60.
-
- Tateno H (2010) SUEL‐related lectins, a lectin family widely distributed throughout organisms. Biosci Biotechnol Biochem 74:1141–1144. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

