Identification and Expression Profiling of the Auxin Response Factors in Dendrobium officinale under Abiotic Stresses
- PMID: 28471373
- PMCID: PMC5454840
- DOI: 10.3390/ijms18050927
Identification and Expression Profiling of the Auxin Response Factors in Dendrobium officinale under Abiotic Stresses
Abstract
Auxin response factor (ARF) proteins play roles in plant responses to diverse environmental stresses by binding specifically to the auxin response element in the promoters of target genes. Using our latest public Dendrobium transcriptomes, a comprehensive characterization and analysis of 14 DnARF genes were performed. Three selected DnARFs, including DnARF1, DnARF4, and DnARF6, were confirmed to be nuclear proteins according to their transient expression in epidermal cells of Nicotiana benthamiana leaves. Furthermore, the transcription activation abilities of DnARF1, DnARF4, and DnARF6 were tested in a yeast system. Our data showed that DnARF6 is a transcriptional activator in Dendrobium officinale. To uncover the basic information of DnARF gene responses to abiotic stresses, we analyzed their expression patterns under various hormones and abiotic treatments. Based on our data, several hormones and significant stress responsive DnARF genes have been identified. Since auxin and ARF genes have been identified in many plant species, our data is imperative to reveal the function of ARF mediated auxin signaling in the adaptation to the challenging Dendrobium environment.
Keywords: ARF; Dendrobium officinale; abiotic stress; auxin; transcriptional activator.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




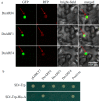
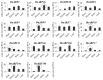

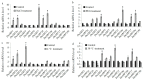
References
-
- Yue R., Tie S., Sun T., Zhang L., Yang Y., Qi J., Yan S., Han X., Wang H., Shen C. Genome-wide identification and expression profiling analysis of ZmPIN, ZmPILS, ZmLAX and ZmABCB auxin transporter gene families in maize (Zea mays L.) under various abiotic stresses. PLoS ONE. 2015;10:e0118751. doi: 10.1371/journal.pone.0118751. - DOI - PMC - PubMed
-
- Shen C., Yue R., Yang Y., Zhang L., Sun T., Xu L., Tie S., Wang H. Genome-wide identification and expression profiling analysis of the AUX/IAA gene family in Medicago truncatula during the early phase of sinorhizobium meliloti infection. PLoS ONE. 2014;9:e107495. doi: 10.1371/journal.pone.0107495. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

