Whole-Genome Sequencing Accurately Identifies Resistance to Extended-Spectrum β-Lactams for Major Gram-Negative Bacterial Pathogens
- PMID: 28472260
- PMCID: PMC5850535
- DOI: 10.1093/cid/cix417
Whole-Genome Sequencing Accurately Identifies Resistance to Extended-Spectrum β-Lactams for Major Gram-Negative Bacterial Pathogens
Abstract
Background: There is marked interest in using DNA-based methods to detect antimicrobial resistance (AMR), with targeted polymerase chain reaction (PCR) approaches increasingly being incorporated into clinical care. Whole-genome sequencing (WGS) could offer significant advantages over targeted PCR for AMR detection, particularly for species where mutations are major drivers of AMR.
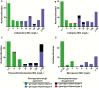
Methods: Illumina MiSeq WGS and broth microdilution (BMD) assays were performed on 90 bloodstream isolates of the 4 most common gram-negative bacteria causing bloodstream infections in neutropenic patients. The WGS data, including both gene presence/absence and detection of mutations in an array of AMR-relevant genes, were used to predict resistance to 4 β-lactams commonly used in the empiric treatment of neutropenic fever. The genotypic predictions were then compared to phenotypic resistance as determined by BMD and by commercial methods during routine patient care.
Results: Of 133 putative instances of resistance to the β-lactams of interest identified by WGS, only 87 (65%) would have been detected by a typical PCR-based approach. The sensitivity, specificity, and positive and negative predictive values for WGS in predicting AMR were 0.87, 0.98, 0.97, and 0.91, respectively. Using BMD as the gold standard, our genotypic resistance prediction approach had a significantly higher positive predictive value compared to minimum inhibitory concentrations generated by commercial methods (0.97 vs 0.92; P = .025).
Conclusions: These data demonstrate the potential feasibility of using WGS to guide antibiotic treatment decisions for patients with life-threatening infections for an array of medically important pathogens.
Keywords: antimicrobial resistance; bacteremia; gram-negative bacteria; neutropenic fever; whole-genome sequencing.
© The Author 2017. Published by Oxford University Press for the Infectious Diseases Society of America. All rights reserved. For permissions, e-mail: journals.permissions@oup.com.
Figures



References
-
- Ward C, Stocker K, Begum J, Wade P, Ebrahimsa U, Goldenberg SD. Performance evaluation of the Verigene (Nanosphere) and FilmArray (BioFire) molecular assays for identification of causative organisms in bacterial bloodstream infections. Eur J Clin Microbiol Infect Dis 2015; 34:487–96. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

