Annotating function to differentially expressed LincRNAs in myelodysplastic syndrome using a network-based method
- PMID: 28472271
- PMCID: PMC5860060
- DOI: 10.1093/bioinformatics/btx280
Annotating function to differentially expressed LincRNAs in myelodysplastic syndrome using a network-based method
Abstract
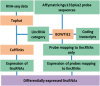
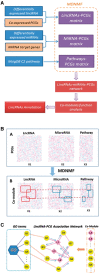
Motivation: Long non-coding RNAs (lncRNAs) have been implicated in the regulation of diverse biological functions. The number of newly identified lncRNAs has increased dramatically in recent years but their expression and function have not yet been described from most diseases. To elucidate lncRNA function in human disease, we have developed a novel network based method (NLCFA) integrating correlations between lncRNA, protein coding genes and noncoding miRNAs. We have also integrated target gene associations and protein-protein interactions and designed our model to provide information on the combined influence of mRNAs, lncRNAs and miRNAs on cellular signal transduction networks.
Results: We have generated lncRNA expression profiles from the CD34+ haematopoietic stem and progenitor cells (HSPCs) from patients with Myelodysplastic syndromes (MDS) and healthy donors. We report, for the first time, aberrantly expressed lncRNAs in MDS and further prioritize biologically relevant lncRNAs using the NLCFA. Taken together, our data suggests that aberrant levels of specific lncRNAs are intimately involved in network modules that control multiple cancer-associated signalling pathways and cellular processes. Importantly, our method can be applied to prioritize aberrantly expressed lncRNAs for functional validation in other diseases and biological contexts.
Availability and implementation: The method is implemented in R language and Matlab.
Contact: xizhou@wakehealth.edu.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author (2017). Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com
Figures


Similar articles
-
Integrated Analysis of Long Non-Coding RNA and mRNA Expression Profile in Myelodysplastic Syndromes.Clin Lab. 2020 May 1;66(5). doi: 10.7754/Clin.Lab.2019.190939. Clin Lab. 2020. PMID: 32390397
-
Full high-throughput sequencing analysis of differences in expression profiles of long noncoding RNAs and their mechanisms of action in systemic lupus erythematosus.Arthritis Res Ther. 2019 Mar 5;21(1):70. doi: 10.1186/s13075-019-1853-7. Arthritis Res Ther. 2019. PMID: 30836987 Free PMC article.
-
An integrative transcriptomic analysis reveals p53 regulated miRNA, mRNA, and lncRNA networks in nasopharyngeal carcinoma.Tumour Biol. 2016 Mar;37(3):3683-95. doi: 10.1007/s13277-015-4156-x. Epub 2015 Oct 13. Tumour Biol. 2016. PMID: 26462838
-
Gene regulation by long non-coding RNAs and its biological functions.Nat Rev Mol Cell Biol. 2021 Feb;22(2):96-118. doi: 10.1038/s41580-020-00315-9. Epub 2020 Dec 22. Nat Rev Mol Cell Biol. 2021. PMID: 33353982 Free PMC article. Review.
-
PltDB: a blood platelets-based gene expression database for disease investigation.Bioinformatics. 2022 May 26;38(11):3143-3145. doi: 10.1093/bioinformatics/btac278. Bioinformatics. 2022. PMID: 35438150 Review.
Cited by
-
Deciphering the Non-Coding RNA Landscape of Pediatric Acute Myeloid Leukemia.Cancers (Basel). 2022 Apr 22;14(9):2098. doi: 10.3390/cancers14092098. Cancers (Basel). 2022. PMID: 35565228 Free PMC article.
-
An Integrated Regulatory Network Based on Comprehensive Analysis of mRNA Expression, Gene Methylation and Expression of Long Non-coding RNAs (lncRNAs) in Myelodysplastic Syndromes.Front Oncol. 2019 Mar 29;9:200. doi: 10.3389/fonc.2019.00200. eCollection 2019. Front Oncol. 2019. PMID: 30984623 Free PMC article.
-
LncRNA Profiling Reveals That the Deregulation of H19, WT1-AS, TCL6, and LEF1-AS1 Is Associated with Higher-Risk Myelodysplastic Syndrome.Cancers (Basel). 2020 Sep 23;12(10):2726. doi: 10.3390/cancers12102726. Cancers (Basel). 2020. PMID: 32977510 Free PMC article.
-
Systematical Identification of Breast Cancer-Related Circular RNA Modules for Deciphering circRNA Functions Based on the Non-Negative Matrix Factorization Algorithm.Int J Mol Sci. 2019 Feb 20;20(4):919. doi: 10.3390/ijms20040919. Int J Mol Sci. 2019. PMID: 30791568 Free PMC article.
-
Hypoplastic myelodysplastic syndrome and acquired aplastic anemia: Immune‑mediated bone marrow failure syndromes (Review).Int J Oncol. 2022 Jan;60(1):7. doi: 10.3892/ijo.2021.5297. Epub 2021 Dec 27. Int J Oncol. 2022. PMID: 34958107 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

