BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes
- PMID: 28472356
- PMCID: PMC5570230
- DOI: 10.1093/nar/gkx346
BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes
Abstract
Antibodies have become an indispensable tool for many biotechnological and clinical applications. They bind their molecular target (antigen) by recognizing a portion of its structure (epitope) in a highly specific manner. The ability to predict epitopes from antigen sequences alone is a complex task. Despite substantial effort, limited advancement has been achieved over the last decade in the accuracy of epitope prediction methods, especially for those that rely on the sequence of the antigen only. Here, we present BepiPred-2.0 (http://www.cbs.dtu.dk/services/BepiPred/), a web server for predicting B-cell epitopes from antigen sequences. BepiPred-2.0 is based on a random forest algorithm trained on epitopes annotated from antibody-antigen protein structures. This new method was found to outperform other available tools for sequence-based epitope prediction both on epitope data derived from solved 3D structures, and on a large collection of linear epitopes downloaded from the IEDB database. The method displays results in a user-friendly and informative way, both for computer-savvy and non-expert users. We believe that BepiPred-2.0 will be a valuable tool for the bioinformatics and immunology community.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
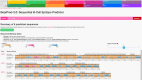
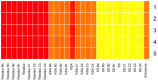
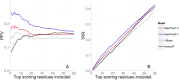
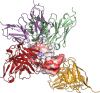
References
-
- Shirai H., Prades C., Vita R., Marcatili P., Popovic B., Xu J., Overington J.P., Hirayama K., Soga S., Tsunoyama K. et al. . Antibody informatics for drug discovery. Biochim. Biophys. Acta. 2014; 1844:2002–2015. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

