WIG1 is crucial for AGO2-mediated ACOT7 mRNA silencing via miRNA-dependent and -independent mechanisms
- PMID: 28472401
- PMCID: PMC5499809
- DOI: 10.1093/nar/gkx307
WIG1 is crucial for AGO2-mediated ACOT7 mRNA silencing via miRNA-dependent and -independent mechanisms
Abstract
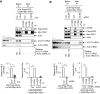
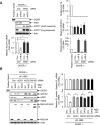
RNA-binding proteins (RBPs) are involved in mRNA splicing, maturation, transport, translation, storage and turnover. Here, we identified ACOT7 mRNA as a novel target of human WIG1. ACOT7 mRNA decay was triggered by the microRNA miR-9 in a WIG1-dependent manner via classic recruitment of Argonaute 2 (AGO2). Interestingly, AGO2 was also recruited to ACOT7 mRNA in a WIG1-dependent manner in the absence of miR-9, which indicates an alternative model whereby WIG1 controls AGO2-mediated gene silencing. The WIG1-AGO2 complex attenuated translation initiation via an interaction with translation initiation factor 5B (eIF5B). These results were confirmed using a WIG1 tethering system based on the MS2 bacteriophage coat protein and a reporter construct containing an MS2-binding site, and by immunoprecipitation of WIG1 and detection of WIG1-associated proteins using liquid chromatography-tandem mass spectrometry. We also identified WIG1-binding motifs using photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation analyses. Altogether, our data indicate that WIG1 governs the miRNA-dependent and the miRNA-independent recruitment of AGO2 to lower the stability of and suppress the translation of ACOT7 mRNA.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures






Similar articles
-
eIF5B regulates the expression of PD-L1 in prostate cancer cells by interacting with Wig1.BMC Cancer. 2021 Sep 15;21(1):1022. doi: 10.1186/s12885-021-08749-w. BMC Cancer. 2021. PMID: 34525951 Free PMC article.
-
Wig1 prevents cellular senescence by regulating p21 mRNA decay through control of RISC recruitment.EMBO J. 2012 Nov 14;31(22):4289-303. doi: 10.1038/emboj.2012.286. Epub 2012 Oct 19. EMBO J. 2012. PMID: 23085987 Free PMC article.
-
Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54.PLoS Biol. 2006 Jul;4(7):e210. doi: 10.1371/journal.pbio.0040210. PLoS Biol. 2006. PMID: 16756390 Free PMC article.
-
[Regulation of hepatitis C virus genome replication by microRNA-122].Uirusu. 2015;65(2):277-286. doi: 10.2222/jsv.65.277. Uirusu. 2015. PMID: 27760927 Review. Japanese.
-
miRNPs: versatile regulators of gene expression in vertebrate cells.Biochem Soc Trans. 2009 Oct;37(Pt 5):931-5. doi: 10.1042/BST0370931. Biochem Soc Trans. 2009. PMID: 19754429 Review.
Cited by
-
AGO2 Mediates MYC mRNA Stability in Hepatocellular Carcinoma.Mol Cancer Res. 2020 Apr;18(4):612-622. doi: 10.1158/1541-7786.MCR-19-0805. Epub 2020 Jan 15. Mol Cancer Res. 2020. PMID: 31941754 Free PMC article.
-
PTBP3 splicing factor promotes hepatocellular carcinoma by destroying the splicing balance of NEAT1 and pre-miR-612.Oncogene. 2018 Dec;37(50):6399-6413. doi: 10.1038/s41388-018-0416-8. Epub 2018 Aug 1. Oncogene. 2018. Retraction in: Oncogene. 2023 Apr;42(15):1263. doi: 10.1038/s41388-023-02648-z. PMID: 30068940 Retracted.
-
RNA-Binding Proteins in Cancer: Functional and Therapeutic Perspectives.Cancers (Basel). 2020 Sep 21;12(9):2699. doi: 10.3390/cancers12092699. Cancers (Basel). 2020. PMID: 32967226 Free PMC article. Review.
-
eIF5B regulates the expression of PD-L1 in prostate cancer cells by interacting with Wig1.BMC Cancer. 2021 Sep 15;21(1):1022. doi: 10.1186/s12885-021-08749-w. BMC Cancer. 2021. PMID: 34525951 Free PMC article.
-
A novel long noncoding RNA Linc-ASEN represses cellular senescence through multileveled reduction of p21 expression.Cell Death Differ. 2020 Jun;27(6):1844-1861. doi: 10.1038/s41418-019-0467-6. Epub 2019 Dec 9. Cell Death Differ. 2020. PMID: 31819156 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

